[English] 日本語
 Yorodumi
Yorodumi- EMDB-17211: Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RESPIRATORY SUPERCOMPLEX / MEMBRANE PROTEIN / ACTINOBACTERIA / ELECTRON TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationaerobic electron transport chain / cytochrome-c oxidase / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / superoxide dismutase / superoxide dismutase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / ATP synthesis coupled electron transport / respiratory electron transport chain ...aerobic electron transport chain / cytochrome-c oxidase / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / superoxide dismutase / superoxide dismutase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / ATP synthesis coupled electron transport / respiratory electron transport chain / monooxygenase activity / electron transport chain / 2 iron, 2 sulfur cluster binding / iron ion binding / copper ion binding / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.3 Å | |||||||||
 Authors Authors | Kovalova T / Krol S / Sjostrand D / Riepl D / Gamiz-Hernandez A / Brzezinski P / Kaila V / Hogbom M | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Long-range charge transfer mechanism of the IIIIV mycobacterial supercomplex. Authors: Daniel Riepl / Ana P Gamiz-Hernandez / Terezia Kovalova / Sylwia M Król / Sophie L Mader / Dan Sjöstrand / Martin Högbom / Peter Brzezinski / Ville R I Kaila /  Abstract: Aerobic life is powered by membrane-bound redox enzymes that shuttle electrons to oxygen and transfer protons across a biological membrane. Structural studies suggest that these energy-transducing ...Aerobic life is powered by membrane-bound redox enzymes that shuttle electrons to oxygen and transfer protons across a biological membrane. Structural studies suggest that these energy-transducing enzymes operate as higher-order supercomplexes, but their functional role remains poorly understood and highly debated. Here we resolve the functional dynamics of the 0.7 MDa IIIIV obligate supercomplex from Mycobacterium smegmatis, a close relative of M. tuberculosis, the causative agent of tuberculosis. By combining computational, biochemical, and high-resolution (2.3 Å) cryo-electron microscopy experiments, we show how the mycobacterial supercomplex catalyses long-range charge transport from its menaquinol oxidation site to the binuclear active site for oxygen reduction. Our data reveal proton and electron pathways responsible for the charge transfer reactions, mechanistic principles of the quinone catalysis, and how unique molecular adaptations, water molecules, and lipid interactions enable the proton-coupled electron transfer (PCET) reactions. Our combined findings provide a mechanistic blueprint of mycobacterial supercomplexes and a basis for developing drugs against pathogenic bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17211.map.gz emd_17211.map.gz | 567.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17211-v30.xml emd-17211-v30.xml emd-17211.xml emd-17211.xml | 41.4 KB 41.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17211.png emd_17211.png | 85.4 KB | ||
| Masks |  emd_17211_msk_1.map emd_17211_msk_1.map | 600.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17211.cif.gz emd-17211.cif.gz | 10.4 KB | ||
| Others |  emd_17211_half_map_1.map.gz emd_17211_half_map_1.map.gz emd_17211_half_map_2.map.gz emd_17211_half_map_2.map.gz | 556.7 MB 556.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17211 http://ftp.pdbj.org/pub/emdb/structures/EMD-17211 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17211 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17211 | HTTPS FTP |
-Related structure data
| Related structure data |  8ovdMC  8ovcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17211.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17211.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
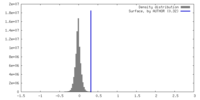
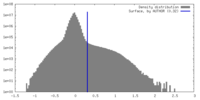
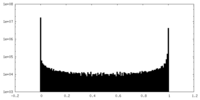
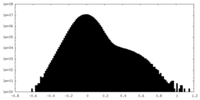
| Density |
| ||||||||||||||||||||||||||||||||||||
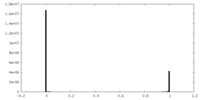
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17211_msk_1.map emd_17211_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
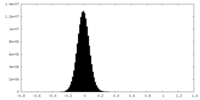
| Density Histograms |
-Half map: #2
| File | emd_17211_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
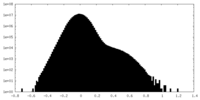
| Density Histograms |
-Half map: #1
| File | emd_17211_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
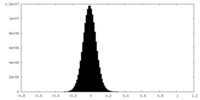
| Density Histograms |
- Sample components
Sample components
+Entire : The respiratory supercomplex
+Supramolecule #1: The respiratory supercomplex
+Macromolecule #1: Cytochrome bc1 complex cytochrome c subunit
+Macromolecule #2: Cytochrome bc1 complex cytochrome c subunit
+Macromolecule #3: Cytochrome bc1 complex cytochrome b subunit
+Macromolecule #4: Transmembrane protein
+Macromolecule #5: Probable cytochrome c oxidase subunit 3
+Macromolecule #6: Cytochrome c oxidase polypeptide 4
+Macromolecule #7: Cytochrome c oxidase subunit 1
+Macromolecule #8: cytochrome-c oxidase
+Macromolecule #9: Cytochrome c oxidase subunit
+Macromolecule #10: Uncharacterized protein MSMEG_4692/MSMEI_4575
+Macromolecule #11: LpqE protein
+Macromolecule #12: Superoxide dismutase [Cu-Zn]
+Macromolecule #13: HEME C
+Macromolecule #14: MENAQUINONE-9
+Macromolecule #15: acyl-phosphatidyl-myo-inositol dimannoside (AcPIM2)
+Macromolecule #16: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #17: [(2~{R})-3-[[(1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-[(2~{R},3~{S...
+Macromolecule #18: (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3...
+Macromolecule #19: (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate
+Macromolecule #20: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #21: CARDIOLIPIN
+Macromolecule #22: TRIDECANE
+Macromolecule #23: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #24: HEME-A
+Macromolecule #25: COPPER (II) ION
+Macromolecule #26: MAGNESIUM ION
+Macromolecule #27: CALCIUM ION
+Macromolecule #28: (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate
+Macromolecule #29: PALMITIC ACID
+Macromolecule #30: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)