-Search query
-Search result
Showing 1 - 50 of 59 items for (author: jensen & pe)

EMDB-16321: 
Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG

PDB-8by6: 
Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG

EMDB-17763: 
Structure of the human nuclear cap-binding complex bound to ARS2[147-871] and m7GTP

EMDB-17784: 
Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA

PDB-8pmp: 
Structure of the human nuclear cap-binding complex bound to ARS2[147-871] and m7GTP

PDB-8pnt: 
Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA

EMDB-14441: 
E. coli C-P lyase bound to a PhnK/PhnL dual ABC dimer and ADP + Pi
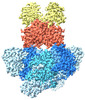
EMDB-14442: 
E. coli C-P lyase bound to PhnK/PhnL dual ABC dimer with AMPPNP and PhnK E171Q mutation

PDB-7z15: 
E. coli C-P lyase bound to a PhnK/PhnL dual ABC dimer and ADP + Pi

PDB-7z16: 
E. coli C-P lyase bound to PhnK/PhnL dual ABC dimer with AMPPNP and PhnK E171Q mutation

EMDB-14443: 
E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation

EMDB-14444: 
E. coli C-P lyase bound to a PhnK ABC dimer and ATP

EMDB-14445: 
E. coli C-P lyase bound to a single PhnK ABC domain

PDB-7z17: 
E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation

PDB-7z18: 
E. coli C-P lyase bound to a PhnK ABC dimer and ATP

PDB-7z19: 
E. coli C-P lyase bound to a single PhnK ABC domain
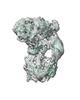
EMDB-13847: 
Cryo-EM structure of native human A2ML1

EMDB-13848: 
Structure of TEV cleaved A2ML1 (A2ML1-TE)
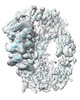
EMDB-13849: 
Structure of TEV conjugated A2ML1 (A2ML1-TC)
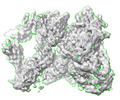
EMDB-13850: 
Structure of TEV cleaved A2ML1 dimer (A2ML1-TT dimer)

PDB-7q5z: 
Cryo-EM structure of native human A2ML1

PDB-7q60: 
Structure of TEV cleaved A2ML1 (A2ML1-TE)

PDB-7q61: 
Structure of TEV conjugated A2ML1 (A2ML1-TC)

PDB-7q62: 
Structure of TEV cleaved A2ML1 dimer (A2ML1-TT dimer)

EMDB-13364: 
UVC treated Human apoferritin

EMDB-13402: 
Cryo-EM map of UVC-treated ICP1 Bacteriophage capsid

EMDB-13403: 
Cryo-EM map of WT ICP1 bacteriophage capsid

PDB-7pf1: 
UVC treated Human apoferritin

EMDB-23060: 
The stress-sensing domain of activated IRE1a forms helical filaments in narrow ER membrane tubes

EMDB-23058: 
The stress-sensing domain of activated IRE1a forms helical filaments in narrow ER membrane tubes

EMDB-22897: 
Disulfide Stabilized Norovirus GI.1 VLP Shell Region

PDB-7kjp: 
Disulfide Stabilized Norovirus GI.1 VLP Shell Region

EMDB-20712: 
In vivo structure of the Legionella type II secretion system by electron cryotomography (aligning the IM-complex).

EMDB-20713: 
In vivo structure of the Legionella type II secretion system by electron cryotomography (aligning the OM-complex).

EMDB-0142: 
Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers.

PDB-6h5s: 
Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers.

EMDB-0141: 
Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers.

PDB-6h5q: 
Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers.

EMDB-0225: 
Mouse serotonin 5-HT3 receptor, serotonin-bound, F conformation

EMDB-0226: 
Mouse serotonin 5-HT3 receptor, serotonin-bound, I1 conformation

EMDB-0227: 
Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation

EMDB-0228: 
Mouse serotonin 5-HT3 receptor, tropisetron-bound, T conformation

PDB-6hin: 
Mouse serotonin 5-HT3 receptor, serotonin-bound, F conformation

PDB-6hio: 
Mouse serotonin 5-HT3 receptor, serotonin-bound, I1 conformation

PDB-6hiq: 
Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation

PDB-6his: 
Mouse serotonin 5-HT3 receptor, tropisetron-bound, T conformation

EMDB-3398: 
Electron Cryotomography and subvolume averaging of the cytoplasmic chemoreceptor array in Vibrio cholerae

EMDB-2414: 
Signaling in Chemoreceptor Arrays Through Mobility Control of Kinase Domains - kinase activity state locked off - tsr mutant M222R

EMDB-5541: 
Signaling in Chemoreceptor Arrays Through Mobility Control of Kinase Domains - kinase activity state locked off - tsr mutant A413T

EMDB-5542: 
Signaling in Chemoreceptor Arrays Through Mobility Control of Kinase Domains - low kinase activity state - tsr mutant P221D
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
