+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22897 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
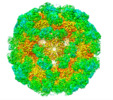
| Title | Disulfide Stabilized Norovirus GI.1 VLP Shell Region | |||||||||||||||
 Map data Map data | Sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Norovirus / Stabilized / VLP / Shell / VIRUS LIKE PARTICLE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationT=3 icosahedral viral capsid / host cell cytoplasm / identical protein binding Similarity search - Function | |||||||||||||||
| Biological species |  Norwalk virus (strain GI/Human/United States/Norwalk/1968) / Norwalk virus (strain GI/Human/United States/Norwalk/1968) /  Norovirus Hu/1968/US Norovirus Hu/1968/US | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.86 Å | |||||||||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: NPJ Vaccines / Year: 2020 Journal: NPJ Vaccines / Year: 2020Title: Disulfide stabilization of human norovirus GI.1 virus-like particles focuses immune response toward blockade epitopes. Authors: Raffaello Verardi / Lisa C Lindesmith / Yaroslav Tsybovsky / Jason Gorman / Gwo-Yu Chuang / Caitlin E Edwards / Paul D Brewer-Jensen / Michael L Mallory / Li Ou / Arne Schön / Wei Shi / Ena ...Authors: Raffaello Verardi / Lisa C Lindesmith / Yaroslav Tsybovsky / Jason Gorman / Gwo-Yu Chuang / Caitlin E Edwards / Paul D Brewer-Jensen / Michael L Mallory / Li Ou / Arne Schön / Wei Shi / Ena S Tully / George Georgiou / Ralph S Baric / Peter D Kwong /  Abstract: Human noroviruses are non-enveloped, single-strand RNA viruses that cause pandemic outbreaks of acute gastroenteritis. A bivalent vaccine containing GI.1 and GII.4 virus-like particles (VLPs) has ...Human noroviruses are non-enveloped, single-strand RNA viruses that cause pandemic outbreaks of acute gastroenteritis. A bivalent vaccine containing GI.1 and GII.4 virus-like particles (VLPs) has been shown to be safe and highly immunogenic, but its efficacy and durability have been limited. Here, we show that norovirus GI.1 VLPs are unstable and contain a substantial fraction of dissociated VLP components. Broadly reactive, non-neutralizing antibodies isolated from vaccinated donors bound to the dissociated components, but not to the intact VLPs. Engineering of interprotomer disulfide bonds within the shell domain prevented disassembly of the VLPs, while preserving antibody accessibility to blockade epitopes. Without adjuvant, mice immunized with stabilized GI.1 VLPs developed faster blockade antibody titers compared to immunization with wild-type GI.1 VLPs. In addition, immunization with stabilized particles focused immune responses toward surface-exposed epitopes and away from occluded epitopes. Overall, disulfide-stabilized norovirus GI.1 VLPs elicited improved responses over the non-disulfide-stabilized version, suggesting their promise as candidate vaccines. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22897.map.gz emd_22897.map.gz | 772.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22897-v30.xml emd-22897-v30.xml emd-22897.xml emd-22897.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22897.png emd_22897.png | 313.3 KB | ||
| Masks |  emd_22897_msk_1.map emd_22897_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22897.cif.gz emd-22897.cif.gz | 6.3 KB | ||
| Others |  emd_22897_additional_1.map.gz emd_22897_additional_1.map.gz emd_22897_half_map_1.map.gz emd_22897_half_map_1.map.gz emd_22897_half_map_2.map.gz emd_22897_half_map_2.map.gz | 170.5 MB 755.4 MB 756.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22897 http://ftp.pdbj.org/pub/emdb/structures/EMD-22897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22897 | HTTPS FTP |
-Related structure data
| Related structure data |  7kjpMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22897.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22897.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07325 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22897_msk_1.map emd_22897_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
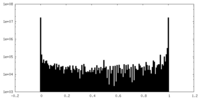
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_22897_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
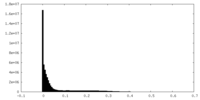
| Density Histograms |
-Half map: #2
| File | emd_22897_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
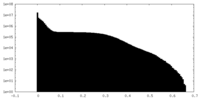
| Density Histograms |
-Half map: #1
| File | emd_22897_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
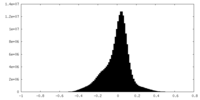
| Density Histograms |
- Sample components
Sample components
-Entire : Norovirus Hu/1968/US
| Entire | Name:  Norovirus Hu/1968/US Norovirus Hu/1968/US |
|---|---|
| Components |
|
-Supramolecule #1: Norovirus Hu/1968/US
| Supramolecule | Name: Norovirus Hu/1968/US / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 524364 / Sci species name: Norovirus Hu/1968/US / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Norwalk virus (strain GI/Human/United States/Norwalk/1968) Norwalk virus (strain GI/Human/United States/Norwalk/1968)Strain: GI/Human/United States/Norwalk/1968 |
| Molecular weight | Theoretical: 56.664961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMMASKDATS SVDGASGAGQ LVPEVNASDP LAMDPVAGSS TAVATAGQVN PIDPWIINNF VQAPQGEFTI SPNNTPGDVL FDLSLGPHL NPFLLHLSQM YNGWVGNMRV RIMLAGCAFT AGKIIVSCIP PGFGSHNLTI AQATLFPHVI ADVRTLDPIE V PLEDVRNV ...String: MMMASKDATS SVDGASGAGQ LVPEVNASDP LAMDPVAGSS TAVATAGQVN PIDPWIINNF VQAPQGEFTI SPNNTPGDVL FDLSLGPHL NPFLLHLSQM YNGWVGNMRV RIMLAGCAFT AGKIIVSCIP PGFGSHNLTI AQATLFPHVI ADVRTLDPIE V PLEDVRNV LFHNNDRNQQ TMRLVCMLYT PLRTCGGTGD SFVVAGRVMT CPSPDFNFLF LVPPTVEQKT RPFTLPNLPL SS LSNSRAP LPISSMGISP DNVQSVQFQN GRCTLDGRLV GTTPVSLSHV AKIRGTSNGT VINLTELDGT PFHPFEGPAP IGF PDLGGC DWHINMTQFG HSSQTQYDVD TTPDTFVPHL GSIQANGIGS GNYVGVLSWI SPPSHPSGSQ VDLWKIPNYG SSIT EATHL APSVYPPGFG EVLVFFMSKM PGPGAYNLPC LLPQEYISHL ASEQAPTVGE AALLHYVDPD TGRNLGEFKA YPDGF LTCV PNGASSGPQQ LPINGVFVFV SWVSRFYQLK PVGTASSARG RLGLRR UniProtKB: Capsid protein VP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Name: PBS |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 777 / Average electron dose: 70.48 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)