[English] 日本語
 Yorodumi
Yorodumi- EMDB-14443: E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
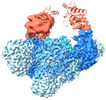
| Title | E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | protein complex / transferase / ABC / hydrolase / lyase / carbon phosphorus | |||||||||
| Function / homology |  Function and homology information Function and homology informationalpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase / alpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase activity / alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase / alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase activity / alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase complex / carbon phosphorus lyase complex / organic phosphonate metabolic process / organic phosphonate transport / organic phosphonate catabolic process / peptide transport ...alpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase / alpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase activity / alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase / alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase activity / alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase complex / carbon phosphorus lyase complex / organic phosphonate metabolic process / organic phosphonate transport / organic phosphonate catabolic process / peptide transport / 4 iron, 4 sulfur cluster binding / lyase activity / protein homodimerization activity / ATP hydrolysis activity / ATP binding / metal ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
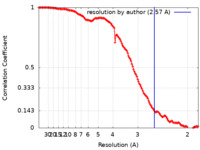
| Method | single particle reconstruction / cryo EM / Resolution: 2.57 Å | |||||||||
 Authors Authors | Amstrup SK / Sofos N | |||||||||
| Funding support |  Denmark, 1 items Denmark, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase. Authors: Søren K Amstrup / Sui Ching Ong / Nicholas Sofos / Jesper L Karlsen / Ragnhild B Skjerning / Thomas Boesen / Jan J Enghild / Bjarne Hove-Jensen / Ditlev E Brodersen /  Abstract: In Escherichia coli, the 14-cistron phn operon encoding carbon-phosphorus lyase allows for utilisation of phosphorus from a wide range of stable phosphonate compounds containing a C-P bond. As part ...In Escherichia coli, the 14-cistron phn operon encoding carbon-phosphorus lyase allows for utilisation of phosphorus from a wide range of stable phosphonate compounds containing a C-P bond. As part of a complex, multi-step pathway, the PhnJ subunit was shown to cleave the C-P bond via a radical mechanism, however, the details of the reaction could not immediately be reconciled with the crystal structure of a 220 kDa PhnGHIJ C-P lyase core complex, leaving a significant gap in our understanding of phosphonate breakdown in bacteria. Here, we show using single-particle cryogenic electron microscopy that PhnJ mediates binding of a double dimer of the ATP-binding cassette proteins, PhnK and PhnL, to the core complex. ATP hydrolysis induces drastic structural remodelling leading to opening of the core complex and reconfiguration of a metal-binding and putative active site located at the interface between the PhnI and PhnJ subunits. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase Authors: Amstrup SK / Sofos N / Karlsen JL / Skjerning RB / Boesen T / Enghild JJ / Hove-Jensen B / Brodersen DE | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14443.map.gz emd_14443.map.gz | 110.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14443-v30.xml emd-14443-v30.xml emd-14443.xml emd-14443.xml | 28.6 KB 28.6 KB | Display Display |  EMDB header EMDB header |
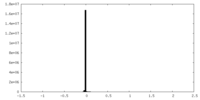
| FSC (resolution estimation) |  emd_14443_fsc.xml emd_14443_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_14443.png emd_14443.png | 174.4 KB | ||
| Filedesc metadata |  emd-14443.cif.gz emd-14443.cif.gz | 7.7 KB | ||
| Others |  emd_14443_additional_1.map.gz emd_14443_additional_1.map.gz emd_14443_half_map_1.map.gz emd_14443_half_map_1.map.gz emd_14443_half_map_2.map.gz emd_14443_half_map_2.map.gz | 4.7 MB 203.9 MB 203.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14443 http://ftp.pdbj.org/pub/emdb/structures/EMD-14443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14443 | HTTPS FTP |
-Related structure data
| Related structure data |  7z17MC  7z15C  7z16C  7z18C  7z19C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14443.map.gz / Format: CCP4 / Size: 219.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14443.map.gz / Format: CCP4 / Size: 219.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92189 Å | ||||||||||||||||||||||||||||||||||||
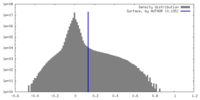
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Map filtered to local resolution
| File | emd_14443_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map filtered to local resolution | ||||||||||||
| Projections & Slices |
| ||||||||||||
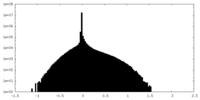
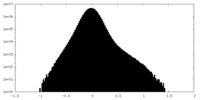
| Density Histograms |
-Half map: Half map B
| File | emd_14443_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
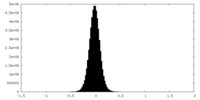
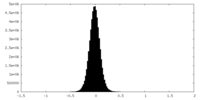
| Density Histograms |
-Half map: Half map A
| File | emd_14443_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
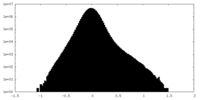
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation
| Entire | Name: E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation |
|---|---|
| Components |
|
-Supramolecule #1: E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation
| Supramolecule | Name: E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: Post ATP hydrolysis by the ATP binding cassette (ABC) protein PhnK |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 277 KDa |
-Macromolecule #1: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subuni...
| Macromolecule | Name: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.545666 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHADTATRQH WMSVLAHSQP AELAARLNAL NITADYEVIR AAETGLVQIQ ARMGGTGERF FAGDATLTRA AVRLTDGTLG YSWVLGRDK QHAERCALID ALMQQSRHFQ NLSETLIAPL DADRMARIAA RQAEVNASRV DFFTMVRGDN A UniProtKB: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG |
-Macromolecule #2: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subuni...
| Macromolecule | Name: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO EC number: alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.045207 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTLETAFMLP VQDAQHSFRR LLKAMSEPGV IVALHQLKRG WQPLNIATTS VLLTLADNDT PVWLSTPLNN DIVNQSLRFH TNAPLVSQP EQATFAVTDE AISSEQLNAL STGTAVAPEA GATLILQVAS LSGGRMLRLT GAGIAEERMI APQLPECILH E LTERPHPF ...String: MTLETAFMLP VQDAQHSFRR LLKAMSEPGV IVALHQLKRG WQPLNIATTS VLLTLADNDT PVWLSTPLNN DIVNQSLRFH TNAPLVSQP EQATFAVTDE AISSEQLNAL STGTAVAPEA GATLILQVAS LSGGRMLRLT GAGIAEERMI APQLPECILH E LTERPHPF PLGIDLILTC GERLLAIPRT THVEVC UniProtKB: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH |
-Macromolecule #3: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subuni...
| Macromolecule | Name: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO EC number: alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.953742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYVAVKGGEK AIDAAHALQE SRRRGDTDLP ELSVAQIEQQ LNLAVDRVMT EGGIADRELA ALALKQASGD NVEAIFLLRA YRTTLAKLA VSEPLDTTGM RLERRISAVY KDIPGGQLLG PTYDYTHRLL DFTLLANGEA PTLTTADSEQ QPSPHVFSLL A RQGLAKFE ...String: MYVAVKGGEK AIDAAHALQE SRRRGDTDLP ELSVAQIEQQ LNLAVDRVMT EGGIADRELA ALALKQASGD NVEAIFLLRA YRTTLAKLA VSEPLDTTGM RLERRISAVY KDIPGGQLLG PTYDYTHRLL DFTLLANGEA PTLTTADSEQ QPSPHVFSLL A RQGLAKFE EDSGAQPDDI TRTPPVYPCS RSSRLQQLMR GDEGYLLALA YSTQRGYGRN HPFAGEIRSG YIDVSIVPEE LG FAVNVGE LLMTECEMVN GFIDPPDEPP HFTRGYGLVF GMSERKAMAM ALVDRALQAP EYGEHATGPA QDEEFVLAHA DNV EAAGFV SHLKLPHYVD FQAELELLKR LQQEKNHG UniProtKB: Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI |
-Macromolecule #4: Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase
| Macromolecule | Name: Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO EC number: alpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.893115 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANLSGYNFA YLDEQTKRMI RRAILKAVAI PGYQVPFGGR EMPMPYGWGT GGIQLTASVI GESDVLKVID QGADDTTNAV SIRNFFKRV TGVNTTERTD DATLIQTRHR IPETPLTEDQ IIIFQVPIPE PLRFIEPRET ETRTMHALEE YGVMQVKLYE D IARFGHIA ...String: MANLSGYNFA YLDEQTKRMI RRAILKAVAI PGYQVPFGGR EMPMPYGWGT GGIQLTASVI GESDVLKVID QGADDTTNAV SIRNFFKRV TGVNTTERTD DATLIQTRHR IPETPLTEDQ IIIFQVPIPE PLRFIEPRET ETRTMHALEE YGVMQVKLYE D IARFGHIA TTYAYPVKVN GRYVMDPSPI PKFDNPKMDM MPALQLFGAG REKRIYAVPP FTRVESLDFD DHPFTVQQWD EP CAICGST HSYLDEVVLD DAGNRMFVCS DTDYCRQQSE AKNQ UniProtKB: Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase |
-Macromolecule #5: Putative phosphonates utilization ATP-binding protein PhnK
| Macromolecule | Name: Putative phosphonates utilization ATP-binding protein PhnK type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.995146 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNQPLLSVNN LTHLYAPGKG FSDVSFDLWP GEVLGIVGES GSGKTTLLKS ISARLTPQQG EIHYENRSLY AMSEADRRRL LRTEWGVVH QHPLDGLRRQ VSAGGNIGER LMATGARHYG DIRATAQKWL EEVEIPANRI DDLPTTFSGG MQQRLQIARN L VTHPKLVF ...String: MNQPLLSVNN LTHLYAPGKG FSDVSFDLWP GEVLGIVGES GSGKTTLLKS ISARLTPQQG EIHYENRSLY AMSEADRRRL LRTEWGVVH QHPLDGLRRQ VSAGGNIGER LMATGARHYG DIRATAQKWL EEVEIPANRI DDLPTTFSGG MQQRLQIARN L VTHPKLVF MDEPTGGLDV SVQARLLDLL RGLVVELNLA VVIVTHDLGV ARLLADRLLV MKQGQVVESG LTDRVLDDPH HP YTQLLVS SVLQNENLYF QGQFGSWSHP QFEKGGGSGG GSGGGSWSHP QFEK UniProtKB: Putative phosphonates utilization ATP-binding protein PhnK |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: alpha-D-ribose-1,2-cyclic-phosphate-5-phosphate
| Macromolecule | Name: alpha-D-ribose-1,2-cyclic-phosphate-5-phosphate / type: ligand / ID: 7 / Number of copies: 1 / Formula: I9X |
|---|---|
| Molecular weight | Theoretical: 292.074 Da |
| Chemical component information |  ChemComp-I8Z: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: ATP up from a concentration from 0.25 to 1.5 and MgCl2 to a concentration of 6 mM was added 15 seconds before the sample was plunge frozen | |||||||||||||||||||||
| Grid | Model: UltrAuFoil R0.6/1 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR / Details: 10 mA in an GloQube, Quorum | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 310 K / Instrument: LEICA EM GP / Details: Blotting for 6-9 seconds before plunging. | |||||||||||||||||||||
| Details | Sample was produced under ATP to ADP+Pi turnover conditions. Sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 62.0 e/Å2 Details: Images were collected in movie-mode with a total of 56 frames |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 135000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)