-Search query
-Search result
Showing 1 - 50 of 69 items for (author: huang & yh)

EMDB-38216: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2
EMDB-34866: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-34869: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-35775: 
The rice Na+/H+ antiporter SOS1 in an auto-inhibited state

EMDB-35950: 
The truncated rice Na+/H+ antiporter SOS1 (1-976) in a constitutively active state
EMDB-34870: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome
EMDB-34867: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-34868: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-34863: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-34864: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-34860: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-34861: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-34862: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome

EMDB-34710: 
Cryo-EM structure of WeiTsing

EMDB-35522: 
Cryo-EM structure of the TUG891 bound GPR120-Giq complex(mask on receptor)

EMDB-35523: 
Cryo-EM structure of the TUG891 bound GPR120-Giq complex(mask on Giq-scFV16 complex)

EMDB-35524: 
Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi1 complex(mask on receptor)

EMDB-35525: 
Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi1 complex(mask on Gil-scFV16 complex)

EMDB-35529: 
Cryo-EM structure of the TUG891 bound GPR120-Giq complex (consensus map)

EMDB-35533: 
Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex(consensus map)

EMDB-35356: 
Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex

EMDB-35357: 
Cryo-EM structure of the linoleic acid bound GPR120-Gi complex

EMDB-35358: 
Cryo-EM structure of the oleic acid bound GPR120-Gi complex

EMDB-35359: 
Cryo-EM structure of the TUG891 bound GPR120-Gi complex

EMDB-35360: 
Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex

EMDB-29736: 
Cryo-EM structure of the TUG891 bound GPR120-Giq complex

EMDB-32928: 
Cryo-EM Structure of Arabidopsis CRY2 in active conformation

EMDB-32929: 
Cryo-EM Structure of Arabidopsis CRY2 tetramer in complex with CIB1 fragment

EMDB-31803: 
LolCDE with bound RcsF in nanodiscs
EMDB-32329: 
Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.

EMDB-32332: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.

EMDB-32333: 
Subtomogram averaging of PEDV (Pintung 52) S protein with one protomer in the D0-up conformation and two protomers in the D0-down conformation, determined in situ on intact viral particles

EMDB-32337: 
Subtomogram averaging of PEDV (Pintung 52) S protein with two protomers in the D0-up conformation and one protomer in the D0-down conformation, determined in situ on intact viral particles.

EMDB-32338: 
Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation
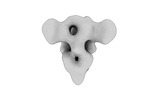
EMDB-32339: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-up conformation determined in situ on intact viral particles.

EMDB-32340: 
Subtomogram averaging of PEDV (Pintung 52) S protein in the postfusion form determined in situ on intact viral particles.

EMDB-33646: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up

EMDB-33647: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up

EMDB-33648: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation
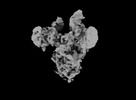
EMDB-33649: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation

EMDB-33700: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-down
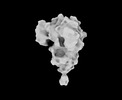
EMDB-33701: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein one D0-up and two D0-down

EMDB-33702: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-up
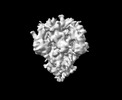
EMDB-33703: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-down

EMDB-33704: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-up and two D0-down
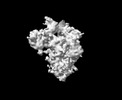
EMDB-33705: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-down and two D0-up
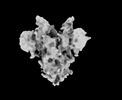
EMDB-33706: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-up

EMDB-32832: 
SARS-CoV-2 Spike in complex with Fab of m31A7

EMDB-32825: 
Negative stain volume of the mono-GlcNAc-decorated SARS-CoV-2 Spike

EMDB-31088: 
LptB2FG-LPS from Klebsiella pneumoniae in nanodiscs
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
