[English] 日本語
 Yorodumi
Yorodumi- EMDB-32338: Cryo-EM map of PEDV S protein with one protomer in the D0-up conf... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | |||||||||||||||||||||
 Map data Map data | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-up conformation. | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | PEDV / Spike / Glycoprotein / VIRAL PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion membrane / membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus | |||||||||||||||||||||
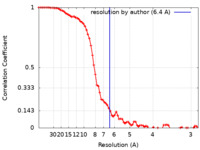
| Method | single particle reconstruction / cryo EM / Resolution: 6.4 Å | |||||||||||||||||||||
 Authors Authors | Hsu STD / Draczkowski P / Wang YS | |||||||||||||||||||||
| Funding support |  Taiwan, 6 items Taiwan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM. Authors: Cheng-Yu Huang / Piotr Draczkowski / Yong-Sheng Wang / Chia-Yu Chang / Yu-Chun Chien / Yun-Han Cheng / Yi-Min Wu / Chun-Hsiung Wang / Yuan-Chih Chang / Yen-Chen Chang / Tzu-Jing Yang / Yu-Xi ...Authors: Cheng-Yu Huang / Piotr Draczkowski / Yong-Sheng Wang / Chia-Yu Chang / Yu-Chun Chien / Yun-Han Cheng / Yi-Min Wu / Chun-Hsiung Wang / Yuan-Chih Chang / Yen-Chen Chang / Tzu-Jing Yang / Yu-Xi Tsai / Kay-Hooi Khoo / Hui-Wen Chang / Shang-Te Danny Hsu /   Abstract: Porcine epidemic diarrhea (PED) is a highly contagious swine disease caused by porcine epidemic diarrhea virus (PEDV). PED causes enteric disorders with an exceptionally high fatality in neonates, ...Porcine epidemic diarrhea (PED) is a highly contagious swine disease caused by porcine epidemic diarrhea virus (PEDV). PED causes enteric disorders with an exceptionally high fatality in neonates, bringing substantial economic losses in the pork industry. The trimeric spike (S) glycoprotein of PEDV is responsible for virus-host recognition, membrane fusion, and is the main target for vaccine development and antigenic analysis. The atomic structures of the recombinant PEDV S proteins of two different strains have been reported, but they reveal distinct N-terminal domain 0 (D0) architectures that may correspond to different functional states. The existence of the D0 is a unique feature of alphacoronavirus. Here we combined cryo-electron tomography (cryo-ET) and cryo-electron microscopy (cryo-EM) to demonstrate in situ the asynchronous S protein D0 motions on intact viral particles of a highly virulent PEDV Pintung 52 strain. We further determined the cryo-EM structure of the recombinant S protein derived from a porcine cell line, which revealed additional domain motions likely associated with receptor binding. By integrating mass spectrometry and cryo-EM, we delineated the complex compositions and spatial distribution of the PEDV S protein N-glycans, and demonstrated the functional role of a key N-glycan in modulating the D0 conformation. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32338.map.gz emd_32338.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32338-v30.xml emd-32338-v30.xml emd-32338.xml emd-32338.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32338_fsc.xml emd_32338_fsc.xml | 7.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_32338.png emd_32338.png | 82.6 KB | ||
| Filedesc metadata |  emd-32338.cif.gz emd-32338.cif.gz | 7.6 KB | ||
| Others |  emd_32338_half_map_1.map.gz emd_32338_half_map_1.map.gz emd_32338_half_map_2.map.gz emd_32338_half_map_2.map.gz | 37.7 MB 37.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32338 http://ftp.pdbj.org/pub/emdb/structures/EMD-32338 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32338 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32338 | HTTPS FTP |
-Related structure data
| Related structure data |  7w73MC  7w6mC  7y6sC  7y6tC  7y6uC  7y6vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32338.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32338.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-up conformation. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM half map (even) of PEDV S protein...
| File | emd_32338_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half map (even) of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-up conformation. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM half map (odd) of PEDV S protein...
| File | emd_32338_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half map (odd) of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-up conformation. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Intact Porcine epidemic diarrhea virus (strain Pintung 52)
| Entire | Name: Intact Porcine epidemic diarrhea virus (strain Pintung 52) |
|---|---|
| Components |
|
-Supramolecule #1: Intact Porcine epidemic diarrhea virus (strain Pintung 52)
| Supramolecule | Name: Intact Porcine epidemic diarrhea virus (strain Pintung 52) type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: The PEDV PT52 virus was propagated in Vero C1008 cells (ATCC No. CRL-1586) and then inactivated by 2% formaldehyde. |
|---|---|
| Source (natural) | Organism:  Porcine epidemic diarrhea virus / Strain: Pintung 52 Porcine epidemic diarrhea virus / Strain: Pintung 52 |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus |
| Molecular weight | Theoretical: 152.005656 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops aethiops (mammal) Chlorocebus aethiops aethiops (mammal) |
| Sequence | String: MKSLTYFWLF LPVLSTLSLP QDVTRCSAKT NFRRFFSKFN VQAPAVVVLG GYLPIGENQG VNSTWYCAGQ HPTASGVHGI FVSHIRGGH GFEIGISQEP FDPSGYQLYL HKATNGNTNA TARLRICQFP SIKTLGPTAN NDVTIGRNCL FNKAIPAHMS E HSVVGITW ...String: MKSLTYFWLF LPVLSTLSLP QDVTRCSAKT NFRRFFSKFN VQAPAVVVLG GYLPIGENQG VNSTWYCAGQ HPTASGVHGI FVSHIRGGH GFEIGISQEP FDPSGYQLYL HKATNGNTNA TARLRICQFP SIKTLGPTAN NDVTIGRNCL FNKAIPAHMS E HSVVGITW DNDRVTVFSD KIYYFYFKND WSRVATKCYN SGGCAMQYVY EPTYYMLNVT SAGEDGISYQ PCTANCIGYA AN VFATEPN GHIPEGFSFN NWFLLSNDST LVHGKVVSNQ PLLVNCLLAI PKIYGLGQFF SFNQTIDGVC NGAAVQRAPE ALR FNINDT SVILAEGSIV LHTALGTNFS FVCSNSSNPH LATFAIPLGA TQVPYYCFFK VDTYNSTVYK FLAVLPPTVR EIVI TKYGD VYVNGFGYLH LGLLDAVTIN FTGHGTDDDV SGFWTIASTN FVDALIEVQG TAIQRILYCD DPVSQLKCSQ VAFDL DDGF YPFSSRNLLS HEQPISFVTL PSFNAHSFVN ITVSASFGGH SGANLIASDT TINGFSSFCV DTRQFTISLS YNVTNS YGY VSNSQDSNCP FTLQSVNDYL SFSKFCVSTS LLASACTIDL FGYPEFGSGV KFTSLYFQFT KGELITGTPK PLEGVTD VS FMTLDVCTKY TIYGFKGEGI ITLTNSSFLA GVYYTSDSGQ LLAFKNVTSG AVYSVTPCSF SEQAAYVDDD IVGVISSL S SSTFNSTREL PGFFYHSNDG SNCTEPVLVY SNIGVCKSGS IGYVPSQSGQ VKIAPTVTGN ISIPTNFSMS IRTEYLQLY NTPVSVDCAT YVCNGNSRCK QLLTQYTAAC KTIESALQLS ARLESVEVNS MLTISEEALQ LATISSFNGD GYNFTNVLGV SVYDPARGR VVQKRSFIED LLFNKVVTNG LGTVDEDYKR CSNGRSVADL VCAQYYSGVM VLPGVVDAEK LHMYSASLIG G MVLGGFTA AAALPFSYAV QARLNYLALQ TDVLQRNQQL LAESFNSAIG NITSAFESVK EASSQTSRGL NTVAHALTKV QE VVNSQGA ALTQLTVQLQ HNFQAISSSI DDIYSRLDIL SADVQVDRLI TGRLSALNAF VAQTLTKYTE VQASRKLAQQ KVN ECVKSQ SQRYGFCGGD GEHIFSLVQA APQGLLFLHT VLVPSDFVDV IAIAGLCVND EIALTLREPG LVLFTHELQN HTAT EYFVS SRRMFEPRKP TVSDFVQIES CVVTYVNLTR DQLPDVIPDY IDVNKTRDEI LASLPNRTGP SLPLDVFNAT YLNLT GEIA DLEQRSESLR NTTEELQSLI YNINNTLVDL EWLNRVETYI KWPWWVWLII FIVLIFVVSL LVFCCISTGF CGCFGC CCA CFSGCCRGPR LQPYEVFEKV HVQ UniProtKB: Spike glycoprotein |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 19 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: Blot for 3 seconds before plunging. Force 0. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | The PEDV PT52 virus was propagated in Vero C1008 cells (ATCC No. CRL-1586) and then inactivated by 2% formaldehyde. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3701 / Average electron dose: 55.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7w73: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)