[English] 日本語
 Yorodumi
Yorodumi- EMDB-33704: Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-up a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-up and two D0-down | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |  Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||||||||||||||
 Authors Authors | Hsu STD / Draczkowski P / Wang YS | |||||||||||||||||||||
| Funding support |  Taiwan, 6 items Taiwan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM. Authors: Cheng-Yu Huang / Piotr Draczkowski / Yong-Sheng Wang / Chia-Yu Chang / Yu-Chun Chien / Yun-Han Cheng / Yi-Min Wu / Chun-Hsiung Wang / Yuan-Chih Chang / Yen-Chen Chang / Tzu-Jing Yang / Yu-Xi ...Authors: Cheng-Yu Huang / Piotr Draczkowski / Yong-Sheng Wang / Chia-Yu Chang / Yu-Chun Chien / Yun-Han Cheng / Yi-Min Wu / Chun-Hsiung Wang / Yuan-Chih Chang / Yen-Chen Chang / Tzu-Jing Yang / Yu-Xi Tsai / Kay-Hooi Khoo / Hui-Wen Chang / Shang-Te Danny Hsu /   Abstract: Porcine epidemic diarrhea (PED) is a highly contagious swine disease caused by porcine epidemic diarrhea virus (PEDV). PED causes enteric disorders with an exceptionally high fatality in neonates, ...Porcine epidemic diarrhea (PED) is a highly contagious swine disease caused by porcine epidemic diarrhea virus (PEDV). PED causes enteric disorders with an exceptionally high fatality in neonates, bringing substantial economic losses in the pork industry. The trimeric spike (S) glycoprotein of PEDV is responsible for virus-host recognition, membrane fusion, and is the main target for vaccine development and antigenic analysis. The atomic structures of the recombinant PEDV S proteins of two different strains have been reported, but they reveal distinct N-terminal domain 0 (D0) architectures that may correspond to different functional states. The existence of the D0 is a unique feature of alphacoronavirus. Here we combined cryo-electron tomography (cryo-ET) and cryo-electron microscopy (cryo-EM) to demonstrate in situ the asynchronous S protein D0 motions on intact viral particles of a highly virulent PEDV Pintung 52 strain. We further determined the cryo-EM structure of the recombinant S protein derived from a porcine cell line, which revealed additional domain motions likely associated with receptor binding. By integrating mass spectrometry and cryo-EM, we delineated the complex compositions and spatial distribution of the PEDV S protein N-glycans, and demonstrated the functional role of a key N-glycan in modulating the D0 conformation. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33704.map.gz emd_33704.map.gz | 11.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33704-v30.xml emd-33704-v30.xml emd-33704.xml emd-33704.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33704_fsc.xml emd_33704_fsc.xml | 6 KB | Display |  FSC data file FSC data file |
| Images |  emd_33704.png emd_33704.png | 32.8 KB | ||
| Others |  emd_33704_half_map_1.map.gz emd_33704_half_map_1.map.gz emd_33704_half_map_2.map.gz emd_33704_half_map_2.map.gz | 21.3 MB 21.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33704 http://ftp.pdbj.org/pub/emdb/structures/EMD-33704 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33704 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33704 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33704.map.gz / Format: CCP4 / Size: 23 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33704.map.gz / Format: CCP4 / Size: 23 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33704_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
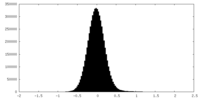
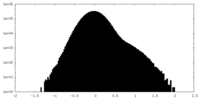
| Density Histograms |
-Half map: #2
| File | emd_33704_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
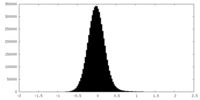
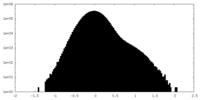
| Density Histograms |
- Sample components
Sample components
-Entire : recombinant PEDV PT52 S protein expressed from HEK293F cells
| Entire | Name: recombinant PEDV PT52 S protein expressed from HEK293F cells |
|---|---|
| Components |
|
-Supramolecule #1: recombinant PEDV PT52 S protein expressed from HEK293F cells
| Supramolecule | Name: recombinant PEDV PT52 S protein expressed from HEK293F cells type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Porcine epidemic diarrhea virus / Strain: Pintung 52 Porcine epidemic diarrhea virus / Strain: Pintung 52 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Macromolecule #1: recombinant PEDV PT52 S T326I expressed from HEK293F cells
| Macromolecule | Name: recombinant PEDV PT52 S T326I expressed from HEK293F cells type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porcine epidemic diarrhea virus / Strain: Pintung52 Porcine epidemic diarrhea virus / Strain: Pintung52 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKSLTYFWLF LPVLSTLSLP QDVTRCSAKT NFRRFFSKFN VQAPAVVVLG GYLPIGENQG VNSTWYCAGQ HPTASGVHGI FVSHIRGGHG FEIGISQEPF DPSGYQLYLH KATNGNTNAT ARLRICQFPS IKTLGPTANN DVTIGRNCLF NKAIPAHMSE HSVVGITWDN ...String: MKSLTYFWLF LPVLSTLSLP QDVTRCSAKT NFRRFFSKFN VQAPAVVVLG GYLPIGENQG VNSTWYCAGQ HPTASGVHGI FVSHIRGGHG FEIGISQEPF DPSGYQLYLH KATNGNTNAT ARLRICQFPS IKTLGPTANN DVTIGRNCLF NKAIPAHMSE HSVVGITWDN DRVTVFSDKI YYFYFKNDWS RVATKCYNSG GCAMQYVYEP TYYMLNVTSA GEDGISYQPC TANCIGYAAN VFATEPNGHI PEGFSFNNWF LLSNDSTLVH GKVVSNQPLL VNCLLAIPKI YGLGQFFSFN QTIDGVCNGA AVQRAPEALR FNINDISVIL AEGSIVLHTA LGTNFSFVCS NSSNPHLATF AIPLGATQVP YYCFFKVDTY NSTVYKFLAV LPPTVREIVI TKYGDVYVNG FGYLHLGLLD AVTINFTGHG TDDDVSGFWT IASTNFVDAL IEVQGTAIQR ILYCDDPVSQ LKCSQVAFDL DDGFYPFSSR NLLSHEQPIS FVTLPSFNAH SFVNITVSAS FGGHSGANLI ASDTTINGFS SFCVDTRQFT ISLSYNVTNS YGYVSNSQDS NCPFTLQSVN DYLSFSKFCV STSLLASACT IDLFGYPEFG SGVKFTSLYF QFTKGELITG TPKPLEGVTD VSFMTLDVCT KYTIYGFKGE GIITLTNSSF LAGVYYTSDS GQLLAFKNVT SGAVYSVTPC SFSEQAAYVD DDIVGVISSL SSSTFNSTRE LPGFFYHSND GSNCTEPVLV YSNIGVCKSG SIGYVPSQSG QVKIAPTVTG NISIPTNFSM SIRTEYLQLY NTPVSVDCAT YVCNGNSRCK QLLTQYTAAC KTIESALQLS ARLESVEVNS MLTISEEALQ LATISSFNGD GYNFTNVLGV SVYDPARGRV VQKRSFIEDL LFNKVVTNGL GTVDEDYKRC SNGRSVADLV CAQYYSGVMV LPGVVDAEKL HMYSASLIGG MVLGGFTAAA ALPFSYAVQA RLNYLALQTD VLQRNQQLLA ESFNSAIGNI TSAFESVKEA SSQTSRGLNT VAHALTKVQE VVNSQGAALT QLTVQLQHNF QAISSSIDDI YSRLDPPSAD VQVDRLITGR LSALNAFVAQ TLTKYTEVQA SRKLAQQKVN ECVKSQSQRY GFCGGDGEHI FSLVQAAPQG LLFLHTVLVP SDFVDVIAIA GLCVNDEIAL TLREPGLVLF THELQNHTAT EYFVSSRRMF EPRKPTVSDF VQIESCVVTY VNLTRDQLPD VIPDYIDVNK TRDEILASLP NRTGPSLPLD VFNATYLNLT GEIADLEQRS ESLRNTTEEL QSLIYNINNT LVDLEWLNRV ETYIKWPEFG SGGYIPEAPR DGQAYVRKDG EWVLLSTFLK GQDNSADIQH SGRPLESRGP FEQKLISEED LNMHTGHHHH HH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Component:
Details: Blot for 3 seconds before plunging. Force 0. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-down |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Details | Data were recorded with stage tilt at 0 and 30 degree |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 2574 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)