+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31088 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | LptB2FG-LPS from Klebsiella pneumoniae in nanodiscs | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | membrane protein / ABC transporter | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationlipopolysaccharide transport / ATP-binding cassette (ABC) transporter complex / transmembrane transport / ATP hydrolysis activity / ATP binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Klebsiella pneumoniae subsp. pneumoniae 1158 (bacteria) / Klebsiella pneumoniae subsp. pneumoniae 1158 (bacteria) /  Klebsiella pneumoniae subsp. pneumoniae (bacteria) Klebsiella pneumoniae subsp. pneumoniae (bacteria) | ||||||||||||
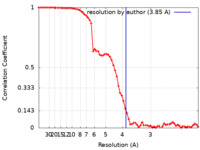
| Method | single particle reconstruction / cryo EM / Resolution: 3.85 Å | ||||||||||||
 Authors Authors | Luo QS / Shi HG / Huang YH | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Biochem Biophys Res Commun / Year: 2021 Journal: Biochem Biophys Res Commun / Year: 2021Title: Cryo-EM structures of LptBFG and LptBFGC from Klebsiella pneumoniae in complex with lipopolysaccharide. Authors: Qingshan Luo / Huigang Shi / Xueqing Xu /  Abstract: Lipopolysaccharide (LPS) is an essential component of the outer membrane (OM) in most Gram-negative bacteria. LPS transport from the inner membrane (IM) to the OM is achieved by seven ...Lipopolysaccharide (LPS) is an essential component of the outer membrane (OM) in most Gram-negative bacteria. LPS transport from the inner membrane (IM) to the OM is achieved by seven lipopolysaccharide transport proteins (LptA-G). LptBFG, an type VI ATP-binding cassette (ABC) transporter, forms a stable complex with LptC, extracts LPS from the IM and powers LPS transport to the OM. Here we report the cryo-EM structures of LptBFG and LptBFGC from Klebsiella pneumoniae in complex with LPS. The KpLptBFG-LPS structure provides detailed interactions between LPS and the transporter, while the KpLptBFGC-LPS structure may represent an intermediate state that the transmembrane helix of LptC has not been fully inserted into the transmembrane domains of LptBFG. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31088.map.gz emd_31088.map.gz | 14 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31088-v30.xml emd-31088-v30.xml emd-31088.xml emd-31088.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31088_fsc.xml emd_31088_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_31088.png emd_31088.png | 106.1 KB | ||
| Masks |  emd_31088_msk_1.map emd_31088_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31088.cif.gz emd-31088.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31088 http://ftp.pdbj.org/pub/emdb/structures/EMD-31088 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31088 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31088 | HTTPS FTP |
-Validation report
| Summary document |  emd_31088_validation.pdf.gz emd_31088_validation.pdf.gz | 583.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31088_full_validation.pdf.gz emd_31088_full_validation.pdf.gz | 583.2 KB | Display | |
| Data in XML |  emd_31088_validation.xml.gz emd_31088_validation.xml.gz | 9.4 KB | Display | |
| Data in CIF |  emd_31088_validation.cif.gz emd_31088_validation.cif.gz | 11.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31088 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31088 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31088 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31088 | HTTPS FTP |
-Related structure data
| Related structure data |  7efoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31088.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31088.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_31088_msk_1.map emd_31088_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : LptB2FG from Klebsiella pneumoniae with bound LPS
| Entire | Name: LptB2FG from Klebsiella pneumoniae with bound LPS |
|---|---|
| Components |
|
-Supramolecule #1: LptB2FG from Klebsiella pneumoniae with bound LPS
| Supramolecule | Name: LptB2FG from Klebsiella pneumoniae with bound LPS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae subsp. pneumoniae 1158 (bacteria) Klebsiella pneumoniae subsp. pneumoniae 1158 (bacteria) |
-Macromolecule #1: Lipopolysaccharide export system ATP-binding protein LptB
| Macromolecule | Name: Lipopolysaccharide export system ATP-binding protein LptB type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae subsp. pneumoniae (bacteria) Klebsiella pneumoniae subsp. pneumoniae (bacteria) |
| Molecular weight | Theoretical: 26.879768 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATLTAKNLA KAYKGRRVVE DVSLTVNSGE IVGLLGPNGA GKTTTFYMVV GIVPRDAGNI IIDDEDISLL PLHARARRGI GYLPQEASI FRRLSVYDNL MAVLQIRDDL TSEQREDRAK ELMEEFHIEH LRDSLGQALS GGERRRVEIA RALAANPKFI L LDEPFAGV ...String: MATLTAKNLA KAYKGRRVVE DVSLTVNSGE IVGLLGPNGA GKTTTFYMVV GIVPRDAGNI IIDDEDISLL PLHARARRGI GYLPQEASI FRRLSVYDNL MAVLQIRDDL TSEQREDRAK ELMEEFHIEH LRDSLGQALS GGERRRVEIA RALAANPKFI L LDEPFAGV DPISVIDIKR IIEHLRDSGL GVLITDHNVR ETLAVCERAY IVSQGHLIAH GTPQQILEDE QVKRVYLGED FR L UniProtKB: Lipopolysaccharide export system ATP-binding protein LptB |
-Macromolecule #2: Lipopolysaccharide export system permease protein LptF
| Macromolecule | Name: Lipopolysaccharide export system permease protein LptF type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae subsp. pneumoniae (bacteria) Klebsiella pneumoniae subsp. pneumoniae (bacteria) |
| Molecular weight | Theoretical: 40.431551 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIIIRYLVRE TLKSQLAILF ILLLIFFCQK LVRILGAAVD GDIPTNLVLS LLGLGIPEMA QLILPLSLFL GLLMTLGKLY TESEITVMH ACGLSKAVLI KAAMILALFT GAVAAVNVMW AGPWSSRHQD EVLAEAKANP GMAALAQGQF QQASDGNAVM F IESVNGNR ...String: MIIIRYLVRE TLKSQLAILF ILLLIFFCQK LVRILGAAVD GDIPTNLVLS LLGLGIPEMA QLILPLSLFL GLLMTLGKLY TESEITVMH ACGLSKAVLI KAAMILALFT GAVAAVNVMW AGPWSSRHQD EVLAEAKANP GMAALAQGQF QQASDGNAVM F IESVNGNR FHDVFLAQLR PKGNARPSVV VADSGELSQQ KDGSQVVTLN KGTRFEGTAM LRDFRITDFN NYQAIIGHQA VS ADPDDTE QMDMRTLWKT HTDRARAELH WRFTLVATVF IMALMVVPLS VVNPRQGRVL SMLPAMLLYL VFFLLQTSIK SNG GKGKMD PAIWMWAINL LYFALAVLLN LWDTVPMRRF RARFNKGAA UniProtKB: Lipopolysaccharide export system permease protein LptF |
-Macromolecule #3: LPS export ABC transporter permease LptG
| Macromolecule | Name: LPS export ABC transporter permease LptG / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae subsp. pneumoniae (bacteria) Klebsiella pneumoniae subsp. pneumoniae (bacteria) |
| Molecular weight | Theoretical: 39.662438 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQAFGVLDRY IGKTIFNTIM MTLFMLVSLS GIIKFVDQLK KSGQGSYDAL GAGLYTILSV PKDIQIFFPM AALLGALLGL GMLAQRSEL VVMQASGFTR LQVALAVMKT AIPLVLLTMA IGEWVAPQGE QMARNYRAQQ MYGGSLLSTQ QGLWAKDGHN F VYIERVKG ...String: MQAFGVLDRY IGKTIFNTIM MTLFMLVSLS GIIKFVDQLK KSGQGSYDAL GAGLYTILSV PKDIQIFFPM AALLGALLGL GMLAQRSEL VVMQASGFTR LQVALAVMKT AIPLVLLTMA IGEWVAPQGE QMARNYRAQQ MYGGSLLSTQ QGLWAKDGHN F VYIERVKG NDELGGVSIY AFNPERRLQS VRYAASAKFD SENKVWRLSQ VDESDLTDPK QVTGSQMVSG TWKTNLTPDK LG VVALDPD ALSISGLHNY VKYLKSSGQD PGRYQLNMWS KIFQPLSVAV MMLMALSFIF GPLRSVPMGV RVVTGISFGF IFY VLDQIF GPLTLVYGIP PIIGALLPSA SFFLISLWLM MRKA UniProtKB: Permease |
-Macromolecule #4: (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(...
| Macromolecule | Name: (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl] ...Name: (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid type: ligand / ID: 4 / Number of copies: 1 / Formula: JSG |
|---|---|
| Molecular weight | Theoretical: 2.975178 KDa |
| Chemical component information |  ChemComp-JSG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.4000000000000001 µm |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)