-Search query
-Search result
Showing 1 - 50 of 237 items for (author: hsu & v)

EMDB-37827: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange state

EMDB-37828: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state

EMDB-37829: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate)

EMDB-37830: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution)

PDB-8wt6: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange state

PDB-8wt7: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state

PDB-8wt8: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate)

PDB-8wt9: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution)

EMDB-18334: 
Cryo-EM structure of the inward-facing FLVCR1

EMDB-18335: 
Cryo-EM structure of the inward-facing choline-bound FLVCR1

EMDB-18336: 
Cryo-EM structure of the inward-facing FLVCR2
EMDB-18337: 
Cryo-EM structure of the outward-facing FLVCR2

EMDB-18339: 
Cryo-EM structure of the inward-facing choline-bound FLVCR2

EMDB-19009: 
Cryo-EM structure of the inward-facing ethanolamine-bound FLVCR1

PDB-8qcs: 
Cryo-EM structure of the inward-facing FLVCR1

PDB-8qct: 
Cryo-EM structure of the inward-facing choline-bound FLVCR1

PDB-8qcx: 
Cryo-EM structure of the inward-facing FLVCR2

PDB-8qcy: 
Cryo-EM structure of the outward-facing FLVCR2

PDB-8qd0: 
Cryo-EM structure of the inward-facing choline-bound FLVCR2

PDB-8r8t: 
Cryo-EM structure of the inward-facing ethanolamine-bound FLVCR1

EMDB-40846: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the low position

EMDB-40847: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the high position

EMDB-40848: 
The structure of C-terminal protease CtpA-LbcA complex of Pseudomonas aeruginosa

EMDB-40849: 
Structure of the C-terminal protease CtpA-LbcA complex of Pseudomonas aeruginosa

EMDB-40850: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the high position
EMDB-40851: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the low position

EMDB-40852: 
Structure of the C-terminal protease CtpA-LbcA complex of Pseudomonas aeruginosa

PDB-8sxe: 
Structure of the C-terminal protease CtpA-LbcA complex of Pseudomonas aeruginosa

PDB-8sxf: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the high position

PDB-8sxg: 
The C-terminal protease CtpA-LbcA complex of pseudomonas aeruginosa with the TPR at the low position

PDB-8sxh: 
Structure of the C-terminal protease CtpA-LbcA complex of Pseudomonas aeruginosa
EMDB-29764: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304
EMDB-29765: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs

EMDB-42148: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304

PDB-8g6e: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304

PDB-8g6f: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs

PDB-8ud9: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304

EMDB-29172: 
Cryo-EM structure of Cryptococcus neoformans trehalose-6-phosphate synthase homotetramer in complex with uridine diphosphate and glucose-6-phosphate

PDB-8fhw: 
Cryo-EM structure of Cryptococcus neoformans trehalose-6-phosphate synthase homotetramer in complex with uridine diphosphate and glucose-6-phosphate

EMDB-33942: 
Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2

EMDB-33943: 
Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 1

EMDB-33944: 
Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 4

EMDB-33945: 
Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 3

EMDB-33946: 
Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 2

EMDB-33947: 
Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 1

EMDB-33948: 
Cryo-EM structure of MERS-CoV spike protein, intermediate conformation

EMDB-33949: 
Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation
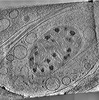
EMDB-29207: 
CryoET tomogram of mitochondria in BACHD mouse model neuron
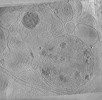
EMDB-29208: 
CryoET tomogram of BACHD mouse model neuron showing sheet aggregates
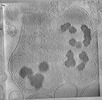
EMDB-29210: 
CryoET tomogram of purified mitochondria from HD patient iPSC-derived neuron (Q109)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
