[English] 日本語
 Yorodumi
Yorodumi- EMDB-33948: Cryo-EM structure of MERS-CoV spike protein, intermediate conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of MERS-CoV spike protein, intermediate conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MERS-CoV / Spike / Glycoprotein / Viral protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationreceptor-mediated endocytosis of virus by host cell / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane ...receptor-mediated endocytosis of virus by host cell / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Human betacoronavirus 2c EMC/2012 Human betacoronavirus 2c EMC/2012 | |||||||||
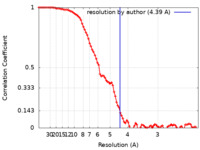
| Method | single particle reconstruction / cryo EM / Resolution: 4.39 Å | |||||||||
 Authors Authors | Hsu STD / Chang NE / Weng ZW / Yang TJ / Draczkowski P | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries. Authors: Yu-Xi Tsai / Ning-En Chang / Klaus Reuter / Hao-Ting Chang / Tzu-Jing Yang / Sören von Bülow / Vidhi Sehrawat / Noémie Zerrouki / Matthieu Tuffery / Michael Gecht / Isabell Louise ...Authors: Yu-Xi Tsai / Ning-En Chang / Klaus Reuter / Hao-Ting Chang / Tzu-Jing Yang / Sören von Bülow / Vidhi Sehrawat / Noémie Zerrouki / Matthieu Tuffery / Michael Gecht / Isabell Louise Grothaus / Lucio Colombi Ciacchi / Yong-Sheng Wang / Min-Feng Hsu / Kay-Hooi Khoo / Gerhard Hummer / Shang-Te Danny Hsu / Cyril Hanus / Mateusz Sikora /      Abstract: Most membrane proteins are modified by covalent addition of complex sugars through N- and O-glycosylation. Unlike proteins, glycans do not typically adopt specific secondary structures and remain ...Most membrane proteins are modified by covalent addition of complex sugars through N- and O-glycosylation. Unlike proteins, glycans do not typically adopt specific secondary structures and remain very mobile, shielding potentially large fractions of protein surface. High glycan conformational freedom hinders complete structural elucidation of glycoproteins. Computer simulations may be used to model glycosylated proteins but require hundreds of thousands of computing hours on supercomputers, thus limiting routine use. Here, we describe GlycoSHIELD, a reductionist method that can be implemented on personal computers to graft realistic ensembles of glycan conformers onto static protein structures in minutes. Using molecular dynamics simulation, small-angle X-ray scattering, cryoelectron microscopy, and mass spectrometry, we show that this open-access toolkit provides enhanced models of glycoprotein structures. Focusing on N-cadherin, human coronavirus spike proteins, and gamma-aminobutyric acid receptors, we show that GlycoSHIELD can shed light on the impact of glycans on the conformation and activity of complex glycoproteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33948.map.gz emd_33948.map.gz | 31.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33948-v30.xml emd-33948-v30.xml emd-33948.xml emd-33948.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33948_fsc.xml emd_33948_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_33948.png emd_33948.png | 25.5 KB | ||
| Filedesc metadata |  emd-33948.cif.gz emd-33948.cif.gz | 6.8 KB | ||
| Others |  emd_33948_additional_1.map.gz emd_33948_additional_1.map.gz emd_33948_half_map_1.map.gz emd_33948_half_map_1.map.gz emd_33948_half_map_2.map.gz emd_33948_half_map_2.map.gz | 59.6 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33948 http://ftp.pdbj.org/pub/emdb/structures/EMD-33948 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33948 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33948 | HTTPS FTP |
-Related structure data
| Related structure data |  7ymzMC  7ymtC  7ymvC  7ymwC  7ymxC  7ymyC  7yn0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33948.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33948.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_33948_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33948_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33948_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : recombinant MERS-CoV (betacoronavirus 2c EMC 2012) fm2P Spike
| Entire | Name: recombinant MERS-CoV (betacoronavirus 2c EMC 2012) fm2P Spike |
|---|---|
| Components |
|
-Supramolecule #1: recombinant MERS-CoV (betacoronavirus 2c EMC 2012) fm2P Spike
| Supramolecule | Name: recombinant MERS-CoV (betacoronavirus 2c EMC 2012) fm2P Spike type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human betacoronavirus 2c EMC/2012 Human betacoronavirus 2c EMC/2012 |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human betacoronavirus 2c EMC/2012 Human betacoronavirus 2c EMC/2012 |
| Molecular weight | Theoretical: 150.654438 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDSWFILVLL GSGLICVSAS YVDVGPDSVK SACIEVDIQQ TFFDKTWPRP IDVSKADGII YPQGRTYSNI TITYQGLFPY QGDHGDMYV YSAGHATGTT PQKLFVANYS QDVKQFANGF VVRIGAAANS TGTVIISPST SATIRKIYPA FMLGSSVGNF S DGKMGRFF ...String: MDSWFILVLL GSGLICVSAS YVDVGPDSVK SACIEVDIQQ TFFDKTWPRP IDVSKADGII YPQGRTYSNI TITYQGLFPY QGDHGDMYV YSAGHATGTT PQKLFVANYS QDVKQFANGF VVRIGAAANS TGTVIISPST SATIRKIYPA FMLGSSVGNF S DGKMGRFF NHTLVLLPDG CGTLLRAFYC ILEPRSGNHC PAGNSYTSFA TYHTPATDCS DGNYNRNASL NSFKEYFNLR NC TFMYTYN ITEDEILEWF GITQTAQGVH LFSSRYVDLY GGNMFQFATL PVYDTIKYYS IIPHSIRSIQ SDRKAWAAFY VYK LQPLTF LLDFSVDGYI RRAIDCGFND LSQLHCSYES FDVESGVYSV SSFEAKPSGS VVEQAEGVEC DFSPLLSGTP PQVY NFKRL VFTNCNYNLT KLLSLFSVND FTCSQISPAA IASNCYSSLI LDYFSYPLSM KSDLSVSSAG PISQFNYKQS FSNPT CLIL ATVPHNLTTI TKPLKYSYIN KCSRLLSDDR TEVPQLVNAN QYSPCVSIVP STVWEDGDYY RKQLSPLEGG GWLVAS GST VAMTEQLQMG FGITVQYGTD TNSVCPKLEF ANDTKIASQL GNCVEYSLYG VSGRGVFQNC TAVGVRQQRF VYDAYQN LV GYYSDDGNYY CLRACVSVPV SVIYDKETKT HATLFGSVAC EHISSTMSQY SRSTRSMLKR RDSTYGPLQT PVGCVLGL V NSSLFVEDCK LPLGQSLCAL PDTPSTLTPA SVGSVPGEMR LASIAFNHPI QVDQLNSSYF KLSIPTNFSF GVTQEYIQT TIQKVTVDCK QYVCNGFQKC EQLLREYGQF CSKINQALHG ANLRQDDSVR NLFASVKSSQ SSPIIPGFGG DFNLTLLEPV SISTGSRSA RSAIEDLLFD KVTIADPGYM QGYDDCMQQG PASARDLICA QYVAGYKVLP PLMDVNMEAA YTSSLLGSIA G VGWTAGLS SFAAIPFAQS IFYRLNGVGI TQQVLSENQK LIANKFNQAL GAMQTGFTTT NEAFQKVQDA VNNNAQALSK LA SELSNTF GAISASIGDI IQRLDPPEQD AQIDRLINGR LTTLNAFVAQ QLVRSESAAL SAQLAKDKVN ECVKAQSKRS GFC GQGTHI VSFVVNAPNG LYFMHVGYYP SNHIEVVSAY GLCDAANPTN CIAPVNGYFI KTNNTRIVDE WSYTGSSFYA PEPI TSLNT KYVAPQVTYQ NISTNLPPPL LGNSTGIDFQ DELDEFFKNV STSIPNFGSL TQINTTLLDL TYEMLSLQQV VKALN ESYI DLKELGNYTY EFGSGGYIPE APRDGQAYVR KDGEWVLLST FLKGQDNSAD IQHSGRPLES RGPFEQKLIS EEDLNM HTG HHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: blot for 2.5 seconds before plunging; blot force: -1; waiting time: 30s.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number grids imaged: 1 / Number real images: 2886 / Average electron dose: 40.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 92000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7ymz: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)