-Search query
-Search result
Showing all 35 items for (author: hirschi & s)

EMDB-16759: 
Cryo-EM structure of retinal-free proteoopsin bound to decanoate

EMDB-16795: 
Cryo-EM structure of pentameric proteorhodopsin A18L mutant

EMDB-16796: 
Cryo-EM structure of hexameric proteorhodopsin A18L mutant

PDB-8cnk: 
Cryo-EM structure of retinal-free proteoopsin bound to decanoate

PDB-8cqc: 
Cryo-EM structure of pentameric proteorhodopsin A18L mutant

PDB-8cqd: 
Cryo-EM structure of hexameric proteorhodopsin A18L mutant

EMDB-29070: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL)

PDB-8ffy: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL)
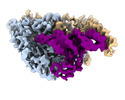
EMDB-26310: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA (GCU)
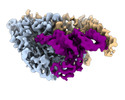
EMDB-26311: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL)

PDB-7u2a: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA (GCU)

PDB-7u2b: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL)

EMDB-24795: 
Mouse heavy chain apoferritin determined in presence of 1.66% glycerol.

EMDB-24796: 
Mouse heavy chain apoferritin determined in presence of 20% glycerol (200keV)

EMDB-24797: 
Mouse heavy chain apoferritin determined in presence of 20% glycerol (300keV)

EMDB-24798: 
Rabbit muscle aldolase determined in presence of 20% v/v glycerol

EMDB-24799: 
Rabbit muscle aldolase determined in the presence of 20% v/v glycerol.

EMDB-11955: 
Cryo-EM structure of the green-light absorbing proteorhodopsin

PDB-7b03: 
Cryo-EM structure of the green-light absorbing proteorhodopsin

EMDB-21516: 
Cryo-EM structure of anti-CRISPR AcrIF9, bound to the type I-F crRNA-guided CRISPR surveillance complex

EMDB-21517: 
Cryo-EM structure of anti-CRISPR AcrIF9 and dsDNA, bound to the type I-F crRNA-guided CRISPR surveillance complex

PDB-6w1x: 
Cryo-EM structure of anti-CRISPR AcrIF9, bound to the type I-F crRNA-guided CRISPR surveillance complex

PDB-6whi: 
Cryo-electron microscopy structure of the type I-F CRISPR RNA-guided surveillance complex bound to the anti-CRISPR AcrIF9

EMDB-9115: 
Structure of the human TRPV3 channel in the apo conformation

EMDB-9117: 
Structure of the human TRPV3 channel in a putative sensitized conformation

EMDB-9119: 
Structure of human TRPV3 in the presence of 2-APB in C4 symmetry

EMDB-9120: 
Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (1)

EMDB-9121: 
Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (2)

PDB-6mho: 
Structure of the human TRPV3 channel in the apo conformation

PDB-6mhs: 
Structure of the human TRPV3 channel in a putative sensitized conformation

PDB-6mhv: 
Structure of human TRPV3 in the presence of 2-APB in C4 symmetry

PDB-6mhw: 
Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (1)

PDB-6mhx: 
Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (2)

EMDB-8764: 
Cryo-electron microscopy structure of a TRPML3 ion channel

PDB-5w3s: 
Cryo-electron microscopy structure of a TRPML3 ion channel
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
