+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9115 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human TRPV3 channel in the apo conformation | ||||||||||||
 Map data Map data | Single-particle cryo-EM reconstruction of human TRPV3 in the apo state | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | membrane protein / ion channel / TRP channel / calcium transport / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of hair cycle / osmosensory signaling pathway / response to temperature stimulus / TRP channels / positive regulation of calcium ion import / sodium channel activity / calcium ion import across plasma membrane / actin filament organization / calcium ion transmembrane transport / calcium channel activity ...negative regulation of hair cycle / osmosensory signaling pathway / response to temperature stimulus / TRP channels / positive regulation of calcium ion import / sodium channel activity / calcium ion import across plasma membrane / actin filament organization / calcium ion transmembrane transport / calcium channel activity / lysosome / receptor complex / cilium / metal ion binding / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Zubcevic L / Herzik MA | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Conformational ensemble of the human TRPV3 ion channel. Authors: Lejla Zubcevic / Mark A Herzik / Mengyu Wu / William F Borschel / Marscha Hirschi / Albert S Song / Gabriel C Lander / Seok-Yong Lee /  Abstract: Transient receptor potential vanilloid channel 3 (TRPV3), a member of the thermosensitive TRP (thermoTRPV) channels, is activated by warm temperatures and serves as a key regulator of normal skin ...Transient receptor potential vanilloid channel 3 (TRPV3), a member of the thermosensitive TRP (thermoTRPV) channels, is activated by warm temperatures and serves as a key regulator of normal skin physiology through the release of pro-inflammatory messengers. Mutations in trpv3 have been identified as the cause of the congenital skin disorder, Olmsted syndrome. Unlike other members of the thermoTRPV channel family, TRPV3 sensitizes upon repeated stimulation, yet a lack of structural information about the channel precludes a molecular-level understanding of TRPV3 sensitization and gating. Here, we present the cryo-electron microscopy structures of apo and sensitized human TRPV3, as well as several structures of TRPV3 in the presence of the common thermoTRPV agonist 2-aminoethoxydiphenyl borate (2-APB). Our results show α-to-π-helix transitions in the S6 during sensitization, and suggest a critical role for the S4-S5 linker π-helix during ligand-dependent gating. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9115.map.gz emd_9115.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9115-v30.xml emd-9115-v30.xml emd-9115.xml emd-9115.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9115.png emd_9115.png | 102 KB | ||
| Filedesc metadata |  emd-9115.cif.gz emd-9115.cif.gz | 6.7 KB | ||
| Others |  emd_9115_half_map_1.map.gz emd_9115_half_map_1.map.gz emd_9115_half_map_2.map.gz emd_9115_half_map_2.map.gz | 45.7 MB 45.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9115 http://ftp.pdbj.org/pub/emdb/structures/EMD-9115 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9115 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9115 | HTTPS FTP |
-Related structure data
| Related structure data |  6mhoMC  9117C  9119C  9120C  9121C  6mhsC  6mhvC  6mhwC  6mhxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9115.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9115.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM reconstruction of human TRPV3 in the apo state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
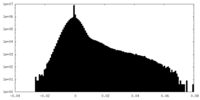
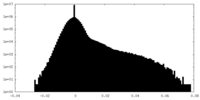
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Odd half map
| File | emd_9115_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Odd half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
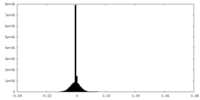
| Density Histograms |
-Half map: Even half map
| File | emd_9115_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Even half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
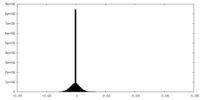
| Density Histograms |
- Sample components
Sample components
-Entire : Human TRPV3 in the apo state
| Entire | Name: Human TRPV3 in the apo state |
|---|---|
| Components |
|
-Supramolecule #1: Human TRPV3 in the apo state
| Supramolecule | Name: Human TRPV3 in the apo state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 3
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 3 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 94.860219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEKAHPKEMV PLMGKRVAAP SGNPAILPEK RPAEITPTKK SAHFFLEIEG FEPNPTVAKT SPPVFSKPMD SNIRQCISGN CDDMDSPQS PQDDVTETPS NPNSPSAQLA KEEQRRKKRR LKKRIFAAVS EGCVEELVEL LVELQELCRR RHDEDVPDFL M HKLTASDT ...String: MEKAHPKEMV PLMGKRVAAP SGNPAILPEK RPAEITPTKK SAHFFLEIEG FEPNPTVAKT SPPVFSKPMD SNIRQCISGN CDDMDSPQS PQDDVTETPS NPNSPSAQLA KEEQRRKKRR LKKRIFAAVS EGCVEELVEL LVELQELCRR RHDEDVPDFL M HKLTASDT GKTCLMKALL NINPNTKEIV RILLAFAEEN DILGRFINAE YTEEAYEGQT ALNIAIERRQ GDIAALLIAA GA DVNAHAK GAFFNPKYQH EGFYFGETPL ALAACTNQPE IVQLLMEHEQ TDITSRDSRG NNILHALVTV AEDFKTQNDF VKR MYDMIL LRSGNWELET TRNNDGLTPL QLAAKMGKAE ILKYILSREI KEKRLRSLSR KFTDWAYGPV SSSLYDLTNV DTTT DNSVL EITVYNTNID NRHEMLTLEP LHTLLHMKWK KFAKHMFFLS FCFYFFYNIT LTLVSYYRPR EEEAIPHPLA LTHKM GWLQ LLGRMFVLIW AMCISVKEGI AIFLLRPSDL QSILSDAWFH FVFFIQAVLV ILSVFLYLFA YKEYLACLVL AMALGW ANM LYYTRGFQSM GMYSVMIQKV ILHDVLKFLF VYIVFLLGFG VALASLIEKC PKDNKDCSSY GSFSDAVLEL FKLTIGL GD LNIQQNSKYP ILFLFLLITY VILTFVLLLN MLIALMGETV ENVSKESERI WRLQRARTIL EFEKMLPEWL RSRFRMGE L CKVAEDDFRL CLRINEVKWT EWKTHVSFLN EDPGPVRRTD FNKIQDSSRN NSKTTLNAFE EVEEFPETSV VDAGLEVLF QGPAAAVDYK DDDDKAHHHH HHHHHH UniProtKB: Transient receptor potential cation channel subfamily V member 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER / Details: Gatan Solarus |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER Details: Sample was manually blotted for 4 seconds prior to plunge-freezing into liquid ethane cooled by liquid nitrogen.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Digitization - Frames/image: 1-48 / Average exposure time: 12.0 sec. / Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 73 / Target criteria: Correlation coefficient |
| Output model |  PDB-6mho: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)