-Search query
-Search result
Showing 1 - 50 of 103 items for (author: hirai & s)

PDB-8wt9: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution)

PDB-8wt8: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate)

PDB-8wt7: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state

PDB-8wt6: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange state

EMDB-37830: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution)

EMDB-37829: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate)

EMDB-37828: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state

EMDB-37827: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange state

EMDB-36920: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-H1147A

EMDB-36921: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146A

EMDB-38247: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146Q

EMDB-38248: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1190A

PDB-8k6j: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-H1147A

PDB-8k6k: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146A

PDB-8xcm: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146Q

PDB-8xcn: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1190A

EMDB-34823: 
Structure of human SGLT2-MAP17 complex with Phlorizin

PDB-8hin: 
Structure of human SGLT2-MAP17 complex with Phlorizin

EMDB-34705: 
Structure of human SGLT2-MAP17 complex with Dapagliflozin

EMDB-34737: 
Structure of human SGLT2-MAP17 complex with Sotagliflozin

PDB-8hez: 
Structure of human SGLT2-MAP17 complex with Dapagliflozin

PDB-8hg7: 
Structure of human SGLT2-MAP17 complex with Sotagliflozin

EMDB-34673: 
Structure of human SGLT2-MAP17 complex with Canagliflozin

PDB-8hdh: 
Structure of human SGLT2-MAP17 complex with Canagliflozin

EMDB-34610: 
Structure of human SGLT2-MAP17 complex with TA1887

PDB-8hb0: 
Structure of human SGLT2-MAP17 complex with TA1887

EMDB-36190: 
Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus

EMDB-36191: 
Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus

PDB-8jej: 
Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus

PDB-8jek: 
Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus

EMDB-37070: 
Cryo-EM structure of the human nucleosome containing H3.8

PDB-8kb5: 
Cryo-EM structure of the human nucleosome containing H3.8

EMDB-34368: 
Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans

EMDB-34369: 
Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans

PDB-8gy2: 
Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans

PDB-8gy3: 
Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans

EMDB-32764: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus

PDB-7wsq: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus

EMDB-32262: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus

PDB-7w2j: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
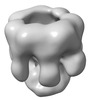
EMDB-32064: 
The oligomerized complex of hemolysin AF3 mutant protomers.
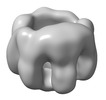
EMDB-32065: 
The oligomerized complex of AG2 hemolysin mutant protomers

EMDB-32591: 
Cryo-EM structure of the nucleosome containing Komagataella pastoris histones

PDB-7wlr: 
Cryo-EM structure of the nucleosome containing Komagataella pastoris histones

EMDB-32151: 
Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1

PDB-7vw6: 
Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1

EMDB-30631: 
The mouse nucleosome structure containing H3mm18

EMDB-31882: 
The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv

PDB-7dbh: 
The mouse nucleosome structure containing H3mm18

PDB-7vbm: 
The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
