[English] 日本語
 Yorodumi
Yorodumi- EMDB-34368: Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Oxidereductase / Membrane-bound protein / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationalcohol dehydrogenase (quinone) / oxidoreductase activity, acting on CH-OH group of donors / outer membrane-bounded periplasmic space / electron transfer activity / iron ion binding / heme binding / calcium ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Gluconobacter oxydans (bacteria) / Gluconobacter oxydans (bacteria) /  Gluconobacter oxydans 621H (bacteria) Gluconobacter oxydans 621H (bacteria) | |||||||||
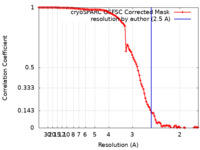
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Adachi T / Miyata T / Makino F / Tanaka H / Namba K / Sowa K / Kitazumi Y / Shirai O | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Acs Catalysis / Year: 2023 Journal: Acs Catalysis / Year: 2023Title: Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases Authors: Adachi T / Miyata T / Makino F / Tanaka H / Namba K / Kano K / Sowa K / Kitazumi Y / Shirai O | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34368.map.gz emd_34368.map.gz | 62.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34368-v30.xml emd-34368-v30.xml emd-34368.xml emd-34368.xml | 27.9 KB 27.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34368_fsc.xml emd_34368_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_34368.png emd_34368.png | 90.9 KB | ||
| Masks |  emd_34368_msk_1.map emd_34368_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34368.cif.gz emd-34368.cif.gz | 7.9 KB | ||
| Others |  emd_34368_half_map_1.map.gz emd_34368_half_map_1.map.gz emd_34368_half_map_2.map.gz emd_34368_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34368 http://ftp.pdbj.org/pub/emdb/structures/EMD-34368 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34368 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34368 | HTTPS FTP |
-Related structure data
| Related structure data |  8gy2MC  8gy3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34368.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34368.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34368_msk_1.map emd_34368_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34368_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34368_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Alcohol dehydrogenase from Gluconobacter oxydans
| Entire | Name: Alcohol dehydrogenase from Gluconobacter oxydans |
|---|---|
| Components |
|
-Supramolecule #1: Alcohol dehydrogenase from Gluconobacter oxydans
| Supramolecule | Name: Alcohol dehydrogenase from Gluconobacter oxydans / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Gluconobacter oxydans (bacteria) Gluconobacter oxydans (bacteria) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Alcohol dehydrogenase (quinone), dehydrogenase subunit
| Macromolecule | Name: Alcohol dehydrogenase (quinone), dehydrogenase subunit type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: alcohol dehydrogenase (quinone) |
|---|---|
| Source (natural) | Organism:  Gluconobacter oxydans 621H (bacteria) / Strain: 621H Gluconobacter oxydans 621H (bacteria) / Strain: 621H |
| Molecular weight | Theoretical: 82.938906 KDa |
| Recombinant expression | Organism:  Gluconobacter oxydans (bacteria) Gluconobacter oxydans (bacteria) |
| Sequence | String: MTSGLLTPIK VTKKRLLSCA AALAFSAAVP VAFAQEDTGT AITSSDNGGH PGDWLSYGRS YSEQRYSPLD QINTENVGKL KLAWHYDLD TNRGQEGTPL IVNGVMYATT NWSKMKALDA ATGKLLWSYD PKVPGNIADR GCCDTVSRGA AYWNGKVYFG T FDGRLIAL ...String: MTSGLLTPIK VTKKRLLSCA AALAFSAAVP VAFAQEDTGT AITSSDNGGH PGDWLSYGRS YSEQRYSPLD QINTENVGKL KLAWHYDLD TNRGQEGTPL IVNGVMYATT NWSKMKALDA ATGKLLWSYD PKVPGNIADR GCCDTVSRGA AYWNGKVYFG T FDGRLIAL DAKTGKLVWS VYTIPKEAQL GHQRSYTVDG APRIAKGKVL IGNGGAEFGA RGFVSAFDAE TGKLDWRFFT VP NPENKPD GAASDDILMS KAYPTWGKNG AWKQQGGGGT VWDSLVYDPV TDLVYLGVGN GSPWNYKFRS EGKGDNLFLG SIV AINPDT GKYVWHFQET PMDEWDYTSV QQIMTLDMPV NGEMRHVIVH APKNGFFYII DAKTGKFITG KPYTYENWAN GLDP VTGRP NYVPDALWTL TGKPWLGIPG ELGGHNFAAM AYSPKTKLVY IPAQQIPLLY DGQKGGFKAY HDAWNLGLDM NKIGL FDDN DPEHVAAKKD FLKVLKGWTV AWDPEKMAPA FTINHKGPWN GGLLATAGNV IFQGLANGEF HAYDATNGND LYSFPA QSA IIAPPVTYTA NGKQYVAVEV GWGGIYPFLY GGVARTSGWT VNHSRVIAFS LDGKDSLPPK NELGFTPVKP VPTYDEA RQ KDGYFMYQTF CSACHGDNAI SGGVLPDLRW SGAPRGRESF YKLVGRGALT AYGMDRFDTS MTPEQIEDIR NFIVKRAN E SYDDEVKARE NSTGVPNDQF LNVPQSTADV PTADHP UniProtKB: Alcohol dehydrogenase (quinone), dehydrogenase subunit |
-Macromolecule #2: Alcohol dehydrogenase (quinone), cytochrome c subunit
| Macromolecule | Name: Alcohol dehydrogenase (quinone), cytochrome c subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: alcohol dehydrogenase (quinone) |
|---|---|
| Source (natural) | Organism:  Gluconobacter oxydans 621H (bacteria) / Strain: 621H Gluconobacter oxydans 621H (bacteria) / Strain: 621H |
| Molecular weight | Theoretical: 51.249598 KDa |
| Recombinant expression | Organism:  Gluconobacter oxydans (bacteria) Gluconobacter oxydans (bacteria) |
| Sequence | String: MLNALTRDRL VSEMKQGWKL AAAIGLMAVS FGAAHAQDAD EALIKRGEYV ARLSDCIACH TALHGQPYAG GLEIKSPIGT IYSTNITPD PEHGIGNYTL EDFTKALRKG IRKDGATVYP AMPYPEFARL SDDDIRAMYA FFMHGVKPVA LQNKAPDISW P LSMRWPLG ...String: MLNALTRDRL VSEMKQGWKL AAAIGLMAVS FGAAHAQDAD EALIKRGEYV ARLSDCIACH TALHGQPYAG GLEIKSPIGT IYSTNITPD PEHGIGNYTL EDFTKALRKG IRKDGATVYP AMPYPEFARL SDDDIRAMYA FFMHGVKPVA LQNKAPDISW P LSMRWPLG MWRAMFVPSM TPGVDKSISD PEVARGEYLV NGPGHCGECH TPRGFGMQVK AYGTAGGNAY LAGGAPIDNW IA PSLRSNS DTGLGRWSED DIVTFLKSGR IDHSAVFGGM ADVVAYSTQH WSDDDLRATA KYLKSMPAVP EGKNLGQDDG QTT ALLNKG GQGNAGAEVY LHNCAICHMN DGTGVNRMFP PLAGNPVVIT DDPTSLANVV AFGGILPPTN SAPSAVAMPG FKNH LSDQE MADVVNFMRK GWGNNAPGTV SASDIQKLRT TGAPVSTAGW NVSSKGWMAY MPQPYGEDWT FSPQTHTGVD DAQ UniProtKB: Alcohol dehydrogenase (quinone), cytochrome c subunit |
-Macromolecule #3: Small subunit of alcohol dehydrogenase
| Macromolecule | Name: Small subunit of alcohol dehydrogenase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: alcohol dehydrogenase (quinone) |
|---|---|
| Source (natural) | Organism:  Gluconobacter oxydans (bacteria) Gluconobacter oxydans (bacteria) |
| Molecular weight | Theoretical: 14.282063 KDa |
| Recombinant expression | Organism:  Gluconobacter oxydans (bacteria) Gluconobacter oxydans (bacteria) |
| Sequence | String: MFRRIVPVLG LALGLGLASQ AAMAQEQSPP PPPAVQGTPG KDFTGVSPAN LAGIMNYCVE QQYVSYDEGN PVLYGLSEKY KATEQTVGN FDYALGTAGY FDSNGKRFYL VAYTNEDDRR AACHAAVKAA QPML UniProtKB: Alcohol dehydrogenase, 15 kDa subunit |
-Macromolecule #4: HEME C
| Macromolecule | Name: HEME C / type: ligand / ID: 4 / Number of copies: 4 / Formula: HEC |
|---|---|
| Molecular weight | Theoretical: 618.503 Da |
| Chemical component information |  ChemComp-HEC: |
-Macromolecule #5: PYRROLOQUINOLINE QUINONE
| Macromolecule | Name: PYRROLOQUINOLINE QUINONE / type: ligand / ID: 5 / Number of copies: 1 / Formula: PQQ |
|---|---|
| Molecular weight | Theoretical: 330.206 Da |
| Chemical component information | 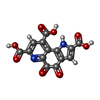 ChemComp-PQQ: |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #7: UBIQUINONE-10
| Macromolecule | Name: UBIQUINONE-10 / type: ligand / ID: 7 / Number of copies: 1 / Formula: U10 |
|---|---|
| Molecular weight | Theoretical: 863.343 Da |
| Chemical component information |  ChemComp-U10: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Temperature | Min: 80.0 K / Max: 80.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.01 mrad |
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3224 / Average exposure time: 3.0 sec. / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 56754 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8gy2: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)