[English] 日本語
 Yorodumi
Yorodumi- EMDB-32151: Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum e... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationformate metabolic process / formate dehydrogenase / formate dehydrogenase (NAD+) activity / oxidoreductase complex / molybdopterin cofactor binding / NADH dehydrogenase activity / NADH dehydrogenase (ubiquinone) activity / respiratory electron transport chain / 2 iron, 2 sulfur cluster binding / FMN binding ...formate metabolic process / formate dehydrogenase / formate dehydrogenase (NAD+) activity / oxidoreductase complex / molybdopterin cofactor binding / NADH dehydrogenase activity / NADH dehydrogenase (ubiquinone) activity / respiratory electron transport chain / 2 iron, 2 sulfur cluster binding / FMN binding / 4 iron, 4 sulfur cluster binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Methylorubrum extorquens AM1 (bacteria) Methylorubrum extorquens AM1 (bacteria) | |||||||||
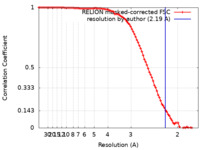
| Method | single particle reconstruction / cryo EM / Resolution: 2.19 Å | |||||||||
 Authors Authors | Yoshikawa T / Makino F / Miyata T / Suzuki Y / Tanaka H / Namba K / Sowa K / Kitazumi Y / Shirai O | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Chem Commun (Camb) / Year: 2022 Journal: Chem Commun (Camb) / Year: 2022Title: Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis. Authors: Tatsushi Yoshikawa / Fumiaki Makino / Tomoko Miyata / Yohei Suzuki / Hideaki Tanaka / Keiichi Namba / Kenji Kano / Keisei Sowa / Yuki Kitazumi / Osamu Shirai /  Abstract: Tungsten-containing formate dehydrogenase from AM1 (FoDH1)-a promising biocatalyst for the interconversion of carbon dioxide/formate and nicotine adenine dinucleotide (NAD)/NADH redox couples-was ...Tungsten-containing formate dehydrogenase from AM1 (FoDH1)-a promising biocatalyst for the interconversion of carbon dioxide/formate and nicotine adenine dinucleotide (NAD)/NADH redox couples-was investigated using structural biology and bioelectrochemistry. FoDH1 is reported to be an enzyme that can realize "direct electron transfer (DET)-type bioelectrocatalysis." However, its 3-D structure, electrode-active sites, and electron transfer (ET) pathways remain unclear. The ET pathways were investigated using structural information, electrostatic interactions between the electrode and the enzyme, and the differences in the substrates. Two electrode-active sites and multiple ET pathways in FoDH1 were discovered. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32151.map.gz emd_32151.map.gz | 60 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32151-v30.xml emd-32151-v30.xml emd-32151.xml emd-32151.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32151_fsc.xml emd_32151_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32151.png emd_32151.png | 71 KB | ||
| Filedesc metadata |  emd-32151.cif.gz emd-32151.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32151 http://ftp.pdbj.org/pub/emdb/structures/EMD-32151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32151 | HTTPS FTP |
-Related structure data
| Related structure data |  7vw6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32151.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32151.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Formate Dehydrogenase 1 from Methylorubrum extorquens AM1
| Entire | Name: Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 |
|---|---|
| Components |
|
-Supramolecule #1: Formate Dehydrogenase 1 from Methylorubrum extorquens AM1
| Supramolecule | Name: Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: Soluble Heterodimer |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens AM1 (bacteria) Methylorubrum extorquens AM1 (bacteria) |
| Molecular weight | Theoretical: 170 KDa |
-Macromolecule #1: Formate dehydrogenase
| Macromolecule | Name: Formate dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: formate dehydrogenase |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens AM1 (bacteria) / Strain: ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1 Methylorubrum extorquens AM1 (bacteria) / Strain: ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1 |
| Molecular weight | Theoretical: 107.464 KDa |
| Sequence | String: MSNGPEPHGN KIEQPEIRAD ERQDAGGPAN GAPSTSGGAY SQGAKSGGQA APDPSGSYGI KDAPVAPATI AFEFDGQQVE AQPGETIWA VAKRLGTHIP HLCHKPDPGY RPDGNCRACM VEIEGERVLA ASCKRTPAIG MKVKSATERA TKARAMVLEL L VADQPERA ...String: MSNGPEPHGN KIEQPEIRAD ERQDAGGPAN GAPSTSGGAY SQGAKSGGQA APDPSGSYGI KDAPVAPATI AFEFDGQQVE AQPGETIWA VAKRLGTHIP HLCHKPDPGY RPDGNCRACM VEIEGERVLA ASCKRTPAIG MKVKSATERA TKARAMVLEL L VADQPERA TSHDPSSHFW VQADVLDVTE SRFPAAERWT SDVSHPAMSV NLDACIQCNL CVRACREVQV NDVIGMAYRA AG SKVVFDF DDPMGGSTCV ACGECVQACP TGALMPAAYL DANQTRTVYP DREVKSLCPY CGVGCQVSYK VKDERIVYAE GVN GPANQN RLCVKGRFGF DYVHHPHRLT VPLIRLENVP KDANDQVDPA NPWTHFREAT WEEALDRAAG GLKAIRDTNG RKAL AGFGS AKGSNEEAYL FQKLVRLGFG TNNVDHCTRL CHASSVAALM EGLNSGAVTA PFSAALDAEV IVVIGANPTV NHPVA ATFL KNAVKQRGAK LIIMDPRRQT LSRHAYRHLA FRPGSDVAML NAMLNVIVTE GLYDEQYIAG YTENFEALRE KIVDFT PEK MASVCGIDAE TLREVARLYA RAKSSLIFWG MGVSQHVHGT DNSRCLIALA LITGQIGRPG TGLHPLRGQN NVQGASD AG LIPMVYPDYQ SVEKDAVREL FEEFWGQSLD PQKGLTVVEI MRAIHAGEIR GMFVEGENPA MSDPDLNHAR HALAMLDH L VVQDLFLTET AFHADVVLPA SAFAEKAGTF TNTDRRVQIA QPVVAPPGDA RQDWWIIQEL ARRLDLDWNY GGPADIFAE MAQVMPSLNN ITWERLEREG AVTYPVDAPD QPGNEIIFYA GFPTESGRAK IVPAAIVPPD EVPDDEFPMV LSTGRVLEHW HTGSMTRRA GVLDALEPEA VAFMAPKELY RLGLRPGGSM RLETRRGAVV LKVRSDRDVP IGMIFMPFCY AEAAANLLTN P ALDPLGKI PEFKFCAARV VPAEAAPMAA E UniProtKB: Tungsten-containing formate dehydrogenase alpha subunit |
-Macromolecule #2: Tungsten-containing formate dehydrogenase beta subunit
| Macromolecule | Name: Tungsten-containing formate dehydrogenase beta subunit type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: formate dehydrogenase |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens AM1 (bacteria) / Strain: ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1 Methylorubrum extorquens AM1 (bacteria) / Strain: ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1 |
| Molecular weight | Theoretical: 62.397508 KDa |
| Sequence | String: MSEASGTVRS FAHPGRGRNV ARAVPKGRQV DPHAKVEIEE LLGTRPRQRD LLIEHLHLIQ DTYGQISADH LAALADEMSL AFAEVFETA TFYAHFDVVK EGEADIPRLT IRVCDSITCA MFGADELLET LQRELASDAV RVVRAPCVGL CDHAPAVEVG H NFLHRADL ...String: MSEASGTVRS FAHPGRGRNV ARAVPKGRQV DPHAKVEIEE LLGTRPRQRD LLIEHLHLIQ DTYGQISADH LAALADEMSL AFAEVFETA TFYAHFDVVK EGEADIPRLT IRVCDSITCA MFGADELLET LQRELASDAV RVVRAPCVGL CDHAPAVEVG H NFLHRADL ASVRAAVEAE DTHAHIPTYV DYDAYRAGGG YATLERLRSG ELPVDDVLKV LDDGGLRGLG GAGFPTGRKW RS VRGEPGP RLMAVNGDEG EPGTFKDQLY LNTDPHRFLE GMLIGAHVVE AADVYIYLRD EYPISREILA REIAKLPEGG TRI HLRRGA GAYICGEESS LIESLEGKRG LPRHKPPFPF QVGLFNRPTL INNIETLFWV RDLIERGAEW WKSHGRNGRV GLRS YSVSG RVKEPGVKLA PAGLTIQELI DEYCGGISDG HSFAAYLPGG ASGGILPASM NDIPLDFGTL EKYGCFIGSA AVVIL SDQD DVRGAALNLM KFFEDESCGQ CTPCRSGTQK ARMLMENGVW DTDLLGELAQ CMRDASICGL GQAASNPVST VIKYFP DLF PEPRAVAAE UniProtKB: Tungsten-containing formate dehydrogenase beta subunit |
-Macromolecule #3: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 3 / Number of copies: 4 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #4: FE2/S2 (INORGANIC) CLUSTER
| Macromolecule | Name: FE2/S2 (INORGANIC) CLUSTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: FES |
|---|---|
| Molecular weight | Theoretical: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-Macromolecule #5: 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,1...
| Macromolecule | Name: 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE type: ligand / ID: 5 / Number of copies: 2 / Formula: MGD |
|---|---|
| Molecular weight | Theoretical: 740.557 Da |
| Chemical component information |  ChemComp-MGD: |
-Macromolecule #6: TUNGSTEN ION
| Macromolecule | Name: TUNGSTEN ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: W |
|---|---|
| Molecular weight | Theoretical: 183.84 Da |
-Macromolecule #7: UNKNOWN ATOM OR ION
| Macromolecule | Name: UNKNOWN ATOM OR ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: UNX |
|---|
-Macromolecule #8: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 8 / Number of copies: 1 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Temperature | Min: 80.0 K / Max: 80.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 7650 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | C2 aperture diameter: 40.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 57471 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7vw6: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)