-Search query
-Search result
Showing 1 - 50 of 1,064 items for (author: gong & a)

EMDB-38695: 
Cryo-EM structure of ATP-DNA-MuB filaments

EMDB-38696: 
CryoEM structure of ADP-DNA-MuB conformation1

EMDB-38697: 
CryoEM structure of ADP-DNA-MuB conformation2

EMDB-38698: 
CryoEM structure of ADP-DNA-MuB conformation3

EMDB-38699: 
CryoEM structure of ADP-DNA-MuB conformation4

PDB-8xvb: 
Cryo-EM structure of ATP-DNA-MuB filaments

PDB-8xvc: 
CryoEM structure of ADP-DNA-MuB conformation1

PDB-8xvd: 
CryoEM structure of ADP-DNA-MuB conformation2
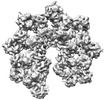
EMDB-38995: 
Structure of the Dark/Dronc complex

PDB-8y6q: 
Structure of the Dark/Dronc complex
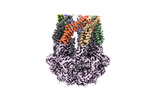
EMDB-36965: 
CryoEM structure of LonC protease hepatmer, apo state
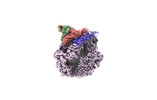
EMDB-36966: 
CryoEM structure of LonC protease open hexamer, apo state
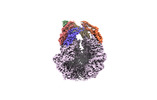
EMDB-36967: 
CryoEM of LonC open pentamer, apo state
EMDB-36968: 
CryoEM structure of LonC heptamer in presence of AGS
EMDB-36969: 
CryoEM structure of LonC protease hexamer in presence of AGS
EMDB-36970: 
CryoEM structure of LonC protease open pentamer in presence of AGS
EMDB-36971: 
CryoEM structure of LonC S582A hepatmer with Lysozyme
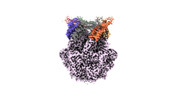
EMDB-36972: 
CryoEM structure of LonC S582A hexamer with Lysozyme
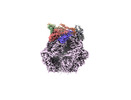
EMDB-36973: 
CryoEM structure of LonC protease S582A open hexamer with lysozyme
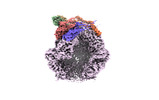
EMDB-36974: 
CryoEM structure of LonC protease S582A open pentamer with lysozyme
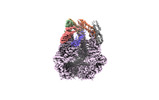
EMDB-36975: 
CryoEM structure of LonC protease open Hexamer, AGS
EMDB-36976: 
CryoEM structure of LonC protease hepatmer with Bortezomib
EMDB-36977: 
CryoEM structure of LonC protease hexamer with Bortezomib

EMDB-36978: 
CryoEM map of LonC protease open hexamer with Bortezomib

EMDB-36979: 
CryoEM map of LonC protease heptamer (heated at 343K)

PDB-8k8v: 
CryoEM structure of LonC protease hepatmer, apo state

PDB-8k8w: 
CryoEM structure of LonC protease open hexamer, apo state

PDB-8k8x: 
CryoEM of LonC open pentamer, apo state

PDB-8k8y: 
CryoEM structure of LonC heptamer in presence of AGS

PDB-8k8z: 
CryoEM structure of LonC protease hexamer in presence of AGS

PDB-8k90: 
CryoEM structure of LonC protease open pentamer in presence of AGS

PDB-8k91: 
CryoEM structure of LonC S582A hepatmer with Lysozyme

PDB-8k92: 
CryoEM structure of LonC S582A hexamer with Lysozyme

PDB-8k93: 
CryoEM structure of LonC protease S582A open hexamer with lysozyme

PDB-8k94: 
CryoEM structure of LonC protease S582A open pentamer with lysozyme

PDB-8k95: 
CryoEM structure of LonC protease open Hexamer, AGS

PDB-8k96: 
CryoEM structure of LonC protease hepatmer with Bortezomib

PDB-8k97: 
CryoEM structure of LonC protease hexamer with Bortezomib

EMDB-43746: 
Plasmodium falciparum 20S proteasome bound to an inhibitor

PDB-8w2f: 
Plasmodium falciparum 20S proteasome bound to an inhibitor
EMDB-35105: 
The cryo-EM structure of human pre-Bact complex

EMDB-35107: 
The cryo-EM structure of human Bact-I complex
EMDB-35108: 
The cryo-EM structure of human Bact-II complex

EMDB-35109: 
The cryo-EM structure of human Bact-III complex

EMDB-35110: 
The cryo-EM structure of human Bact-IV complex
EMDB-35111: 
The cryo-EM structure of human post-Bact complex
EMDB-35113: 
The cryo-EM structure of human C complex

PDB-8i0p: 
The cryo-EM structure of human pre-Bact complex

PDB-8i0r: 
The cryo-EM structure of human Bact-I complex

PDB-8i0s: 
The cryo-EM structure of human Bact-II complex
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
