[English] 日本語
 Yorodumi
Yorodumi- EMDB-36974: CryoEM structure of LonC protease S582A open pentamer with lysozyme -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of LonC protease S582A open pentamer with lysozyme | |||||||||
 Map data Map data | CryoEM map of LonC S582A open pentamer with lysozyme | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lon proteases / chaperone / open pentamer / lysozyme | |||||||||
| Function / homology |  Function and homology information Function and homology informationendopeptidase La / ATP-dependent peptidase activity / protein catabolic process / serine-type endopeptidase activity / proteolysis / ATP binding Similarity search - Function | |||||||||
| Biological species |  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Li M / Hsieh K / Liu H / Zhang S / Gao Y / Gong Q / Zhang K / Chang C / Li S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Fundam Res / Year: 2024 Journal: Fundam Res / Year: 2024Title: Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis Authors: Li M / Liu H / Hsieh KY / Zhang S / Gao Y / Gong Q / Zhang K / Chang CI / Li S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36974.map.gz emd_36974.map.gz | 136.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36974-v30.xml emd-36974-v30.xml emd-36974.xml emd-36974.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36974.png emd_36974.png | 52 KB | ||
| Filedesc metadata |  emd-36974.cif.gz emd-36974.cif.gz | 6.2 KB | ||
| Others |  emd_36974_additional_1.map.gz emd_36974_additional_1.map.gz emd_36974_half_map_1.map.gz emd_36974_half_map_1.map.gz emd_36974_half_map_2.map.gz emd_36974_half_map_2.map.gz | 70 MB 134.2 MB 134.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36974 http://ftp.pdbj.org/pub/emdb/structures/EMD-36974 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36974 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36974 | HTTPS FTP |
-Validation report
| Summary document |  emd_36974_validation.pdf.gz emd_36974_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36974_full_validation.pdf.gz emd_36974_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_36974_validation.xml.gz emd_36974_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_36974_validation.cif.gz emd_36974_validation.cif.gz | 17 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36974 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36974 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36974 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36974 | HTTPS FTP |
-Related structure data
| Related structure data |  8k94MC  8k8vC  8k8wC  8k8xC  8k8yC  8k8zC  8k90C  8k91C  8k92C  8k93C  8k95C  8k96C  8k97C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36974.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36974.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of LonC S582A open pentamer with lysozyme | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
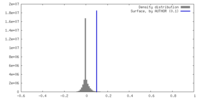
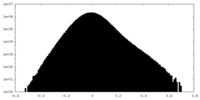
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: CryoEM map of LonC S582A open pentamer with lysozyme
| File | emd_36974_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of LonC S582A open pentamer with lysozyme | ||||||||||||
| Projections & Slices |
| ||||||||||||
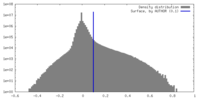
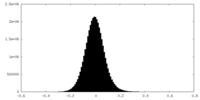
| Density Histograms |
-Half map: CryoEM map of LonC S582A open pentamer with lysozyme
| File | emd_36974_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of LonC S582A open pentamer with lysozyme | ||||||||||||
| Projections & Slices |
| ||||||||||||
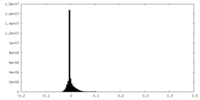
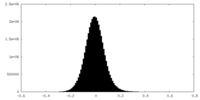
| Density Histograms |
-Half map: CryoEM map of LonC S582A open pentamer with lysozyme
| File | emd_36974_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of LonC S582A open pentamer with lysozyme | ||||||||||||
| Projections & Slices |
| ||||||||||||
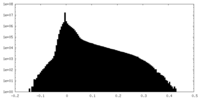
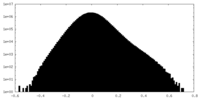
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of LonC protease S582A open pentamer with lysozy...
| Entire | Name: CryoEM structure of LonC protease S582A open pentamer with lysozyme, AGS |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of LonC protease S582A open pentamer with lysozy...
| Supramolecule | Name: CryoEM structure of LonC protease S582A open pentamer with lysozyme, AGS type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Endopeptidase La
| Macromolecule | Name: Endopeptidase La / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) |
| Molecular weight | Theoretical: 80.542352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRLSYEALEW RTPIENSTEP VSLPPPPPFF GQERAREALE LAIRGGFHAY LVGPPSLGKH EALLAYLSTQ SVETPPDLLY VPLSERKVA VLTLPSGQEI HLAEAVEGLL LEVNRLDELF RQGSFLREKT QLEARFKEAR EQQLEALRRE AQEAGFALST N GERLELTG ...String: MRLSYEALEW RTPIENSTEP VSLPPPPPFF GQERAREALE LAIRGGFHAY LVGPPSLGKH EALLAYLSTQ SVETPPDLLY VPLSERKVA VLTLPSGQEI HLAEAVEGLL LEVNRLDELF RQGSFLREKT QLEARFKEAR EQQLEALRRE AQEAGFALST N GERLELTG PGPVPAELSA RLEEVTLGSL AASAELEVAL RRLRRDWALH YLNNRFEPLF QRFPQARAYL EALRARLARY AE TGEPLDP AQWRPNLLTS SSSGTPPPIV YEPYATAPRL FGRLDYLVDR GVWSTNVSLI RPGAVHRAQG GYLILDALSL KRE GTWEAF KRALRNGQVE PVTEPQAPAG LEVEPFPIQM QVILVGTPEA FEGLEEDPAF SELFRIRAEF SPTLPASPEN CTAL GGWLL AQGFQLTQGG LTRLYDEARR MAEQRDRMDA RLVEIRALAE EAAVLGGGLL TAESVEQAIA AREHRSFLSE EEFLR AVQE GVIRLRTTGR AVGEVNSLVV VEAAPYWGRP ARLTARAAPG RDHLISIDRE AGLGGQIFHK AVLTLAGYLR SRYIEH GSL PVTISLAFEQ NYVSIEGDAA GLAELVAALS AIGNLPLRQD LAVTGAVDQT GKVLAVGAIN AKVEGFFRVC KALGLSG TQ GVILPEANLA NLTLRAEVLE AVRAGQFHIY AVETAEQALE ILAGARMEGF RGLQEKIRAG LEAFARLEEG HDKEDREK L AAALEHHHHH H UniProtKB: endopeptidase La |
-Macromolecule #2: Monothiophosphate
| Macromolecule | Name: Monothiophosphate / type: ligand / ID: 2 / Number of copies: 3 / Formula: TS6 |
|---|---|
| Molecular weight | Theoretical: 114.061 Da |
| Chemical component information | 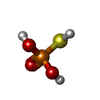 ChemComp-TS6: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)