+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map of LonC protease heptamer (heated at 343K) | |||||||||
 Map data Map data | CryoEM map of LonC protease (Heat at 343 K) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lon proteases / chaperone / hepatmer / high temperature | |||||||||
| Biological species |  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Li M / Hsieh K / Liu H / Zhang S / Gao Y / Gong Q / Zhang K / Chang C / Li S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Fundam Res / Year: 2024 Journal: Fundam Res / Year: 2024Title: Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis Authors: Li M / Liu H / Hsieh KY / Zhang S / Gao Y / Gong Q / Zhang K / Chang CI / Li S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36979.map.gz emd_36979.map.gz | 136.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36979-v30.xml emd-36979-v30.xml emd-36979.xml emd-36979.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36979.png emd_36979.png | 51.8 KB | ||
| Filedesc metadata |  emd-36979.cif.gz emd-36979.cif.gz | 5.2 KB | ||
| Others |  emd_36979_additional_1.map.gz emd_36979_additional_1.map.gz emd_36979_half_map_1.map.gz emd_36979_half_map_1.map.gz emd_36979_half_map_2.map.gz emd_36979_half_map_2.map.gz | 69.6 MB 133.3 MB 133.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36979 http://ftp.pdbj.org/pub/emdb/structures/EMD-36979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36979 | HTTPS FTP |
-Validation report
| Summary document |  emd_36979_validation.pdf.gz emd_36979_validation.pdf.gz | 782.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36979_full_validation.pdf.gz emd_36979_full_validation.pdf.gz | 782.2 KB | Display | |
| Data in XML |  emd_36979_validation.xml.gz emd_36979_validation.xml.gz | 14.6 KB | Display | |
| Data in CIF |  emd_36979_validation.cif.gz emd_36979_validation.cif.gz | 17.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36979 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36979 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36979 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36979 | HTTPS FTP |
-Related structure data
| Related structure data |  8k8vC  8k8wC  8k8xC  8k8yC  8k8zC  8k90C  8k91C  8k92C  8k93C  8k94C  8k95C  8k96C  8k97C C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36979.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36979.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of LonC protease (Heat at 343 K) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||
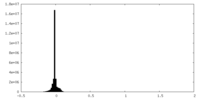
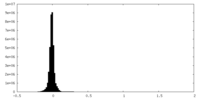
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: CryoEM map of LonC protease (Heat at 343 K)
| File | emd_36979_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of LonC protease (Heat at 343 K) | ||||||||||||
| Projections & Slices |
| ||||||||||||
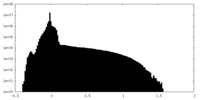
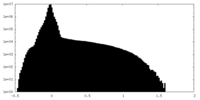
| Density Histograms |
-Half map: CryoEM map of LonC protease (Heat at 343 K)
| File | emd_36979_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of LonC protease (Heat at 343 K) | ||||||||||||
| Projections & Slices |
| ||||||||||||
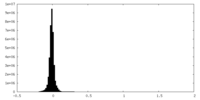
| Density Histograms |
-Half map: CryoEM map of LonC protease (Heat at 343 K)
| File | emd_36979_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of LonC protease (Heat at 343 K) | ||||||||||||
| Projections & Slices |
| ||||||||||||
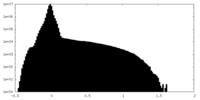
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of LonC protease hepatmer, heated at 343K
| Entire | Name: CryoEM structure of LonC protease hepatmer, heated at 343K |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of LonC protease hepatmer, heated at 343K
| Supramolecule | Name: CryoEM structure of LonC protease hepatmer, heated at 343K type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) |
| Molecular weight | Theoretical: 560 KDa |
-Macromolecule #1: LonC heptamer (heated)
| Macromolecule | Name: LonC heptamer (heated) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MRLSYEALEW RTPIENSTEP VSLPPPPPFF GQERAREALE LAIRGGFHAY LVGPPSLGKH EALLAYLSTQ SVETPPDLLY VPLSERKVAV LTLPSGQEIH LAEAVEGLLL EVNRLDELFR QGSFLREKTQ LEARFKEARE QQLEALRREA QEAGFALSTN GERLELTGPG ...String: MRLSYEALEW RTPIENSTEP VSLPPPPPFF GQERAREALE LAIRGGFHAY LVGPPSLGKH EALLAYLSTQ SVETPPDLLY VPLSERKVAV LTLPSGQEIH LAEAVEGLLL EVNRLDELFR QGSFLREKTQ LEARFKEARE QQLEALRREA QEAGFALSTN GERLELTGPG PVPAELSARL EEVTLGSLAA SAELEVALRR LRRDWALHYL NNRFEPLFQR FPQARAYLEA LRARLARYAE TGEPLDPAQW RPNLLTSSSS GTPPPIVYEP YATAPRLFGR LDYLVDRGVW STNVSLIRPG AVHRAQGGYL ILDALSLKRE GTWEAFKRAL RNGQVEPVTE PQAPAGLEVE PFPIQMQVIL VGTPEAFEGL EEDPAFSELF RIRAEFSPTL PASPENCTAL GGWLLAQGFQ LTQGGLTRLY DEARRMAEQR DRMDARLVEI RALAEEAAVL GGGLLTAESV EQAIAAREHR SFLSEEEFLR AVQEGVIRLR TTGRAVGEVN SLVVVEAAPY WGRPARLTAR AAPGRDHLIS IDREAGLGGQ IFHKAVLTLA GYLRSRYIEH GSLPVTISLA FEQNYVSIEG DSAGLAELVA ALSAIGNLPL RQDLAVTGAV DQTGKVLAVG AINAKVEGFF RVCKALGLSG TQGVILPEAN LANLTLRAEV LEAVRAGQFH IYAVETAEQA LEILAGARME GFRGLQEKIR AGLEAFARLE EGHDKEDREK LAAALEHHHH HH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm |
 Movie
Movie Controller
Controller



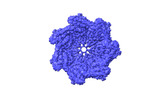














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)