-Search query
-Search result
Showing 1 - 50 of 62 items for (author: gao & gg)

EMDB-34724: 
Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803

PDB-8hfq: 
Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803

EMDB-36342: 
Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist

EMDB-36360: 
cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist

EMDB-36361: 
Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist

EMDB-36426: 
Structure of the adhesion GPCR ADGRL3 in the apo state

PDB-8jj8: 
Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist

PDB-8jjl: 
cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist

PDB-8jjo: 
Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist

PDB-8jmt: 
Structure of the adhesion GPCR ADGRL3 in the apo state

EMDB-33151: 
Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab

PDB-7xdl: 
Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab

EMDB-33130: 
Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with BA7054 and BA7125 fab (local refinement)

EMDB-33140: 
Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with BA7208 and BA7125 fab (local refinement)

EMDB-33142: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab

EMDB-33150: 
Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab

PDB-7xcz: 
Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with BA7054 and BA7125 fab (local refinement)

PDB-7xda: 
Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with BA7208 and BA7125 fab (local refinement)

PDB-7xdb: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab

PDB-7xdk: 
Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab
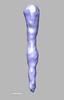
EMDB-28834: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 3
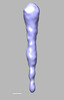
EMDB-28838: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 2

EMDB-28839: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 1
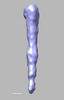
EMDB-28840: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 4
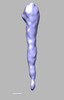
EMDB-28841: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 5
EMDB-28842: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 6

EMDB-28843: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 7

EMDB-28844: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 8

EMDB-33748: 
Spike_GSAS_6P protomer RBD domain bound with R1-32 Fab and ACE2 with 3:3:3 ratio
EMDB-33760: 
Spike_GSAS_6P and R1-32 Fab with 3to1 ratio
EMDB-33764: 
SARS-COV-2 Spike_GSAS_6P bound with two R1-32 Fabs
EMDB-33766: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs

EMDB-33772: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2

PDB-7ydi: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region

PDB-7ydy: 
SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab

PDB-7ye5: 
SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs

PDB-7ye9: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs

PDB-7yeg: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2

EMDB-31058: 
The Csy-AcrIF14 complex

EMDB-31059: 
The Csy-AcrIF14-dsDNA complex

PDB-7ecv: 
The Csy-AcrIF14 complex

PDB-7ecw: 
The Csy-AcrIF14-dsDNA complex

EMDB-31373: 
Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002

EMDB-31381: 
Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120

EMDB-31483: 
Cryo-EM structure of cyanobacterial PBS(delete Lr) from Synechococcus sp.PCC 7002

PDB-7ext: 
Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002

PDB-7eyd: 
Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120

EMDB-30374: 
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs

PDB-7chh: 
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs

EMDB-10462: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
