+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31059 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Csy-AcrIF14-dsDNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inhibitor / complex / IMMUNE SYSTEM / IMMUNE SYSTEM-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationCRISPR-associated protein Csy1 / CRISPR-associated protein (Cas_Csy1) / CRISPR-associated protein Csy2 / CRISPR-associated protein (Cas_Csy2) / CRISPR-associated protein Csy3 / CRISPR-associated protein (Cas_Csy3) Similarity search - Domain/homology | |||||||||
| Biological species |   Moraxella phage Mcat5 (virus) Moraxella phage Mcat5 (virus) | |||||||||
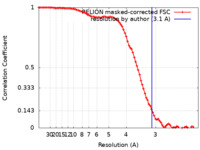
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Zhang LX / Feng Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2021 Journal: Nucleic Acids Res / Year: 2021Title: Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex. Authors: Xi Liu / Laixing Zhang / Yu Xiu / Teng Gao / Ling Huang / Yongchao Xie / Lingguang Yang / Wenhe Wang / Peiyi Wang / Yi Zhang / Maojun Yang / Yue Feng /  Abstract: CRISPR-Cas systems are bacterial adaptive immune systems, and phages counteract these systems using many approaches such as producing anti-CRISPR (Acr) proteins. Here, we report the structures of ...CRISPR-Cas systems are bacterial adaptive immune systems, and phages counteract these systems using many approaches such as producing anti-CRISPR (Acr) proteins. Here, we report the structures of both AcrIF14 and its complex with the crRNA-guided surveillance (Csy) complex. Our study demonstrates that apart from interacting with the Csy complex to block the hybridization of target DNA to the crRNA, AcrIF14 also endows the Csy complex with the ability to interact with non-sequence-specific dsDNA as AcrIF9 does. Further structural studies of the Csy-AcrIF14-dsDNA complex and biochemical studies uncover that the PAM recognition loop of the Cas8f subunit of the Csy complex and electropositive patches within the N-terminal domain of AcrIF14 are essential for the non-sequence-specific dsDNA binding to the Csy-AcrIF14 complex, which is different from the mechanism of AcrIF9. Our findings highlight the prevalence of Acr-induced non-specific DNA binding and shed light on future studies into the mechanisms of such Acr proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31059.map.gz emd_31059.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31059-v30.xml emd-31059-v30.xml emd-31059.xml emd-31059.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31059_fsc.xml emd_31059_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_31059.png emd_31059.png | 99 KB | ||
| Filedesc metadata |  emd-31059.cif.gz emd-31059.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31059 http://ftp.pdbj.org/pub/emdb/structures/EMD-31059 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31059 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31059 | HTTPS FTP |
-Related structure data
| Related structure data |  7ecwMC  7du0C  7ecvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31059.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31059.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : membrane protein
+Supramolecule #1: membrane protein
+Supramolecule #2: membrane protein
+Supramolecule #3: AcrIF14
+Macromolecule #1: Type I-F CRISPR-associated protein Csy1
+Macromolecule #2: CRISPR type I-F/YPEST-associated protein Csy2
+Macromolecule #3: CRISPR-associated protein Csy3
+Macromolecule #4: AcrIF14
+Macromolecule #5: RNA (60-MER)
+Macromolecule #6: 54-MER DNA
+Macromolecule #7: 54-MER DNA
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DARK FIELD |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)