+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31058 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Csy-AcrIF14 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inhibitor / complex / IMMUNE SYSTEM / IMMUNE SYSTEM-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationCRISPR-associated protein Csy1 / CRISPR-associated protein (Cas_Csy1) / CRISPR-associated protein Csy2 / CRISPR-associated protein (Cas_Csy2) / CRISPR-associated protein Csy3 / CRISPR-associated protein (Cas_Csy3) Similarity search - Domain/homology | |||||||||
| Biological species |   Moraxella phage Mcat5 (virus) Moraxella phage Mcat5 (virus) | |||||||||
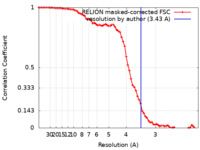
| Method | single particle reconstruction / cryo EM / Resolution: 3.43 Å | |||||||||
 Authors Authors | Zhang LX / Feng Y | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2021 Journal: Nucleic Acids Res / Year: 2021Title: Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex. Authors: Xi Liu / Laixing Zhang / Yu Xiu / Teng Gao / Ling Huang / Yongchao Xie / Lingguang Yang / Wenhe Wang / Peiyi Wang / Yi Zhang / Maojun Yang / Yue Feng /  Abstract: CRISPR-Cas systems are bacterial adaptive immune systems, and phages counteract these systems using many approaches such as producing anti-CRISPR (Acr) proteins. Here, we report the structures of ...CRISPR-Cas systems are bacterial adaptive immune systems, and phages counteract these systems using many approaches such as producing anti-CRISPR (Acr) proteins. Here, we report the structures of both AcrIF14 and its complex with the crRNA-guided surveillance (Csy) complex. Our study demonstrates that apart from interacting with the Csy complex to block the hybridization of target DNA to the crRNA, AcrIF14 also endows the Csy complex with the ability to interact with non-sequence-specific dsDNA as AcrIF9 does. Further structural studies of the Csy-AcrIF14-dsDNA complex and biochemical studies uncover that the PAM recognition loop of the Cas8f subunit of the Csy complex and electropositive patches within the N-terminal domain of AcrIF14 are essential for the non-sequence-specific dsDNA binding to the Csy-AcrIF14 complex, which is different from the mechanism of AcrIF9. Our findings highlight the prevalence of Acr-induced non-specific DNA binding and shed light on future studies into the mechanisms of such Acr proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31058.map.gz emd_31058.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31058-v30.xml emd-31058-v30.xml emd-31058.xml emd-31058.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31058_fsc.xml emd_31058_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_31058.png emd_31058.png | 81.4 KB | ||
| Filedesc metadata |  emd-31058.cif.gz emd-31058.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31058 http://ftp.pdbj.org/pub/emdb/structures/EMD-31058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31058 | HTTPS FTP |
-Related structure data
| Related structure data |  7ecvMC  7du0C  7ecwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31058.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31058.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The Csy-AcrIF14 complex
| Entire | Name: The Csy-AcrIF14 complex |
|---|---|
| Components |
|
-Supramolecule #1: The Csy-AcrIF14 complex
| Supramolecule | Name: The Csy-AcrIF14 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Csy
| Supramolecule | Name: Csy / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4, #6 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: AcrIF14
| Supramolecule | Name: AcrIF14 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:  Moraxella phage Mcat5 (virus) Moraxella phage Mcat5 (virus) |
-Macromolecule #1: Type I-F CRISPR-associated endoribonuclease Cas6/Csy4
| Macromolecule | Name: Type I-F CRISPR-associated endoribonuclease Cas6/Csy4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.399451 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDHYLDIRLR PDPEFPPAQL MSVLFGKLHQ ALVAQGGDRI GVSFPDLDES RSRLGERLRI HASADDLRAL LARPWLEGLR DHLQFGEPA VVPHPTPYRQ VSRVQAKSNP ERLRRRLMRR HDLSEEEARK RIPDTVARAL DLPFVTLRSQ STGQHFRLFI R HGPLQATA EEGGFTCYGL SKGGFVPWF UniProtKB: UNIPROTKB: A0A7G9X1S4 |
-Macromolecule #2: Type I-F CRISPR-associated protein Csy1
| Macromolecule | Name: Type I-F CRISPR-associated protein Csy1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.313254 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLHPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EAVQALSDNA EQAREWRQAF I GITAVKGA ...String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLHPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EAVQALSDNA EQAREWRQAF I GITAVKGA PASHSLAKQL YFPLPGSGYH LLAPLFPTSL VHHVHALLRE ARFGDAAKAA REARSRQESW PHGFSEYPNL AI QKFGGTK PQNISQLNSE RYGENWLLPS LPPHWQRQDQ RAPIRHSSVF EHDFGRSPEV SRLTRTLQRL LAKTRHNNFT IRR YRAQLV GQICDEALQY AARLRELEPG WSATPGCQLH DAEQLWLDPL RAQTDETFLQ RRLRGDWPAE VGNRFANWLN RAVS SDSQI LGSPEAAQWS QELSKELTMF KEILEDERD UniProtKB: Uncharacterized protein |
-Macromolecule #3: CRISPR type I-F/YPEST-associated protein Csy2
| Macromolecule | Name: CRISPR type I-F/YPEST-associated protein Csy2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.244074 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ ...String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ RRKNQRRLTR RLLPGFALVS REALLQQHLE TLRTTLPEAT TLDALLDLCR INFEPPATSS EEEASPPDAA WQ VRDKPGW LVPIPAGYNA LSPLYLPGEV RNARDRETPL RFVENLFGLG EWLSPHRVAA LSDLLWYHHA EPDKGLYRWS TPR FVEHAI A UniProtKB: Uncharacterized protein |
-Macromolecule #4: CRISPR-associated protein Csy3
| Macromolecule | Name: CRISPR-associated protein Csy3 / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.623324 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKPILSTAS VLAFERKLDP SDALMSAGAW AQRDASQEWP AVTVREKSVR GTISNRLKTK DRDPAKLDAS IQSPNLQTVD VANLPSDAD TLKVRFTLRV LGGAGTPSAC NDAAYRDKLL QTVATYVNEQ GFAELARRYA HNLANARFLW RNRVGAEAVE V RINHIRQG ...String: MSKPILSTAS VLAFERKLDP SDALMSAGAW AQRDASQEWP AVTVREKSVR GTISNRLKTK DRDPAKLDAS IQSPNLQTVD VANLPSDAD TLKVRFTLRV LGGAGTPSAC NDAAYRDKLL QTVATYVNEQ GFAELARRYA HNLANARFLW RNRVGAEAVE V RINHIRQG EVARTWRFDA LAIGLRDFKA DAELDALAEL IASGLSGSGH VLLEVVAFAR IGDGQEVFPS QELILDKGDK KG QKSKTLY SVRDAAAIHS QKIGNALRTI DTWYPDEDGL GPIAVEPYGS VTSQGKAYRQ PKQKLDFYTL LDNWVLRDEA PAV EQQHYV IANLIRGGVF GEAEEK UniProtKB: CRISPR-associated protein Csy3 |
-Macromolecule #5: AcrIF14
| Macromolecule | Name: AcrIF14 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Moraxella phage Mcat5 (virus) Moraxella phage Mcat5 (virus) |
| Molecular weight | Theoretical: 14.297373 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKIEMIEIS QNRQNLTAFL HISEIKAINA KLADGVDVDK KSFDEICSIV LEQYQAKQIS NKQASEIFET LAKANKSFKI EKFRCSHGY NEIYKYSPDH EAYLFYCKGG QGQLNKLIAE NGRFM UniProtKB: Tail protein |
-Macromolecule #6: RNA (60-MER)
| Macromolecule | Name: RNA (60-MER) / type: rna / ID: 6 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.265404 KDa |
| Sequence | String: CUAAGAAAUU CACGGCGGGC UUGAUGUCCG CGUCUACCUG GUUCACUGCC GUGUAGGCAG GENBANK: GENBANK: HQ326201.1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DARK FIELD |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)