+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the adhesion GPCR ADGRL3 in the apo state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ADGRL3 / Apo form / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / anchoring junction / electron transport chain / G protein-coupled receptor activity / carbohydrate binding / electron transfer activity / periplasmic space / cell surface receptor signaling pathway / iron ion binding / axon ...: / anchoring junction / electron transport chain / G protein-coupled receptor activity / carbohydrate binding / electron transfer activity / periplasmic space / cell surface receptor signaling pathway / iron ion binding / axon / heme binding / calcium ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.36 Å | |||||||||
 Authors Authors | Tao Y / Guo Q / He B / Zhong Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: A method for structure determination of GPCRs in various states. Authors: Qiong Guo / Binbin He / Yixuan Zhong / Haizhan Jiao / Yinhang Ren / Qinggong Wang / Qiangqiang Ge / Yongxiang Gao / Xiangyu Liu / Yang Du / Hongli Hu / Yuyong Tao /  Abstract: G-protein-coupled receptors (GPCRs) are a class of integral membrane proteins that detect environmental cues and trigger cellular responses. Deciphering the functional states of GPCRs induced by ...G-protein-coupled receptors (GPCRs) are a class of integral membrane proteins that detect environmental cues and trigger cellular responses. Deciphering the functional states of GPCRs induced by various ligands has been one of the primary goals in the field. Here we developed an effective universal method for GPCR cryo-electron microscopy structure determination without the need to prepare GPCR-signaling protein complexes. Using this method, we successfully solved the structures of the β-adrenergic receptor (βAR) bound to antagonistic and agonistic ligands and the adhesion GPCR ADGRL3 in the apo state. For βAR, an intermediate state stabilized by the partial agonist was captured. For ADGRL3, the structure revealed that inactive ADGRL3 adopts a compact fold and that large unusual conformational changes on both the extracellular and intracellular sides are required for activation of adhesion GPCRs. We anticipate that this method will open a new avenue for understanding GPCR structure‒function relationships and drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36426.map.gz emd_36426.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36426-v30.xml emd-36426-v30.xml emd-36426.xml emd-36426.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
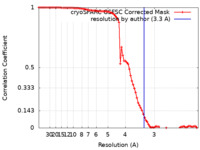
| FSC (resolution estimation) |  emd_36426_fsc.xml emd_36426_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_36426.png emd_36426.png | 41.6 KB | ||
| Filedesc metadata |  emd-36426.cif.gz emd-36426.cif.gz | 6.7 KB | ||
| Others |  emd_36426_half_map_1.map.gz emd_36426_half_map_1.map.gz emd_36426_half_map_2.map.gz emd_36426_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36426 http://ftp.pdbj.org/pub/emdb/structures/EMD-36426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36426 | HTTPS FTP |
-Validation report
| Summary document |  emd_36426_validation.pdf.gz emd_36426_validation.pdf.gz | 672.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36426_full_validation.pdf.gz emd_36426_full_validation.pdf.gz | 672.4 KB | Display | |
| Data in XML |  emd_36426_validation.xml.gz emd_36426_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_36426_validation.cif.gz emd_36426_validation.cif.gz | 20.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36426 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36426 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36426 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36426 | HTTPS FTP |
-Related structure data
| Related structure data |  8jmtMC  8j7eC  8jj8C  8jjlC  8jjoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36426.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36426.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
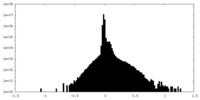
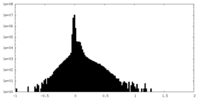
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36426_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
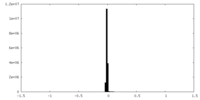
| Density Histograms |
-Half map: #1
| File | emd_36426_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
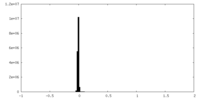
| Density Histograms |
- Sample components
Sample components
-Entire : ADGRL3-mBRIL complex
| Entire | Name: ADGRL3-mBRIL complex |
|---|---|
| Components |
|
-Supramolecule #1: ADGRL3-mBRIL complex
| Supramolecule | Name: ADGRL3-mBRIL complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Adhesion G protein-coupled receptor L3,Soluble cytochrome b562
| Macromolecule | Name: Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 85.439922 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKAV EAREIMWFKT RQGQIAKQPC PAGTIGVSTY LCLAPDGIWD PQGPDLSNCS SPWVNHITQK LKSGETAANI ARELAEQTR NHLNAGDITY SVRAMDQLVG LLDVQLRNLT PGGKDSAARS LNKLQKRERS CRAYVQAMVE TVNNLLQPQA L NAWRDLTT ...String: DYKDDDDKAV EAREIMWFKT RQGQIAKQPC PAGTIGVSTY LCLAPDGIWD PQGPDLSNCS SPWVNHITQK LKSGETAANI ARELAEQTR NHLNAGDITY SVRAMDQLVG LLDVQLRNLT PGGKDSAARS LNKLQKRERS CRAYVQAMVE TVNNLLQPQA L NAWRDLTT SDQLRAATML LHTVEESAFV LADNLLKTDI VRENTDNIKL EVARLSTEGN LEDLKFPENM GHGSTIQLSA NT LKQNGRN GEIRVAFVLY NNLGPYLSTE NASMKLGTEA LSTNHSVIVN SPVITAAINK EFSNKVYLAD PVVFTVKHIK QSE ENFNPN CSFWSYSKRT MTGYWSTQGC RLLTTNKTHT TCSCNHLANF AVLMAHVEVK HSDAVHDLLL DVITWVGILL SLVC LLICI FTFCFFRGLQ SDRNTIHKNL CISLFVAELL FLIGINRTDQ PIACAVFAAL LHFFFLAAFT WMFLEGVQLY IMLVE VFES EHSRRKYFYL VGYGMPALIV AVSAAVDYRS YGTDKVCWLR LDTYFIWSFI GPATLIIMLN VIFLGIALYK MFHHTA DLE DNWETLNDNL KVIEKADNAA QVKDALTKMR AAALDAQKAS GSGSPEMKDF RHGFDILVGQ IDDALKLANE GKVKEAQ AA AEQLKTTRNA YIQKYLERAD NIKSWVIGAI ALLCLLGLTW AFGLMYINES TVIMAYLFTI FNSLQGMFIF IFHCVLQK K VRKEYGKCLR THAASRLEEE LRRRLTEGSH HHHHHHH UniProtKB: Adhesion G protein-coupled receptor L3, Soluble cytochrome b562, Soluble cytochrome b562, Adhesion G protein-coupled receptor L3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)