[English] 日本語
 Yorodumi
Yorodumi- EMDB-36361: Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / Membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of mini excitatory postsynaptic potential / beta2-adrenergic receptor activity / negative regulation of smooth muscle contraction / AMPA selective glutamate receptor signaling pathway / norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure / positive regulation of autophagosome maturation / heat generation / norepinephrine binding / Adrenoceptors / positive regulation of lipophagy ...positive regulation of mini excitatory postsynaptic potential / beta2-adrenergic receptor activity / negative regulation of smooth muscle contraction / AMPA selective glutamate receptor signaling pathway / norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure / positive regulation of autophagosome maturation / heat generation / norepinephrine binding / Adrenoceptors / positive regulation of lipophagy / negative regulation of G protein-coupled receptor signaling pathway / negative regulation of multicellular organism growth / response to psychosocial stress / adrenergic receptor signaling pathway / endosome to lysosome transport / diet induced thermogenesis / positive regulation of cAMP/PKA signal transduction / adenylate cyclase binding / smooth muscle contraction / bone resorption / potassium channel regulator activity / positive regulation of bone mineralization / neuronal dense core vesicle / intercellular bridge / regulation of sodium ion transport / adenylate cyclase-activating adrenergic receptor signaling pathway / brown fat cell differentiation / receptor-mediated endocytosis / response to cold / clathrin-coated endocytic vesicle membrane / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / cellular response to amyloid-beta / mitotic spindle / Cargo recognition for clathrin-mediated endocytosis / amyloid-beta binding / positive regulation of cold-induced thermogenesis / Clathrin-mediated endocytosis / microtubule cytoskeleton / G alpha (s) signalling events / transcription by RNA polymerase II / early endosome / cell surface receptor signaling pathway / lysosome / receptor complex / positive regulation of MAPK cascade / apical plasma membrane / endosome / endosome membrane / Ub-specific processing proteases / cilium / ciliary basal body / protein-containing complex binding / Golgi apparatus / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / identical protein binding / membrane / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
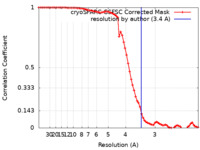
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | He BB / Zhong YX / Guo Q / Tao YY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: A method for structure determination of GPCRs in various states. Authors: Qiong Guo / Binbin He / Yixuan Zhong / Haizhan Jiao / Yinhang Ren / Qinggong Wang / Qiangqiang Ge / Yongxiang Gao / Xiangyu Liu / Yang Du / Hongli Hu / Yuyong Tao /  Abstract: G-protein-coupled receptors (GPCRs) are a class of integral membrane proteins that detect environmental cues and trigger cellular responses. Deciphering the functional states of GPCRs induced by ...G-protein-coupled receptors (GPCRs) are a class of integral membrane proteins that detect environmental cues and trigger cellular responses. Deciphering the functional states of GPCRs induced by various ligands has been one of the primary goals in the field. Here we developed an effective universal method for GPCR cryo-electron microscopy structure determination without the need to prepare GPCR-signaling protein complexes. Using this method, we successfully solved the structures of the β-adrenergic receptor (βAR) bound to antagonistic and agonistic ligands and the adhesion GPCR ADGRL3 in the apo state. For βAR, an intermediate state stabilized by the partial agonist was captured. For ADGRL3, the structure revealed that inactive ADGRL3 adopts a compact fold and that large unusual conformational changes on both the extracellular and intracellular sides are required for activation of adhesion GPCRs. We anticipate that this method will open a new avenue for understanding GPCR structure‒function relationships and drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36361.map.gz emd_36361.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36361-v30.xml emd-36361-v30.xml emd-36361.xml emd-36361.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36361_fsc.xml emd_36361_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_36361.png emd_36361.png | 69.7 KB | ||
| Filedesc metadata |  emd-36361.cif.gz emd-36361.cif.gz | 6.4 KB | ||
| Others |  emd_36361_half_map_1.map.gz emd_36361_half_map_1.map.gz emd_36361_half_map_2.map.gz emd_36361_half_map_2.map.gz | 37.7 MB 37.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36361 http://ftp.pdbj.org/pub/emdb/structures/EMD-36361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36361 | HTTPS FTP |
-Related structure data
| Related structure data |  8jjoMC  8j7eC  8jj8C  8jjlC  8jmtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36361.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36361.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36361_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
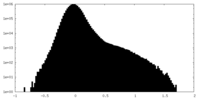
| Projections & Slices |
| ||||||||||||
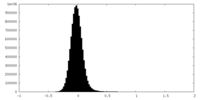
| Density Histograms |
-Half map: #1
| File | emd_36361_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
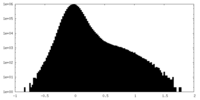
| Projections & Slices |
| ||||||||||||
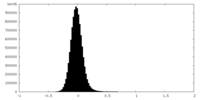
| Density Histograms |
- Sample components
Sample components
-Entire : chimeric beta-2 adrenergic receptor
| Entire | Name: chimeric beta-2 adrenergic receptor |
|---|---|
| Components |
|
-Supramolecule #1: chimeric beta-2 adrenergic receptor
| Supramolecule | Name: chimeric beta-2 adrenergic receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Beta-2 adrenergic receptor,Beta-2 adrenergic receptor,Soluble cyt...
| Macromolecule | Name: Beta-2 adrenergic receptor,Beta-2 adrenergic receptor,Soluble cytochrome b562 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.594102 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKDEVWVV GMGIVMSLIV LAIVFGNVLV ITAIAKFERL QTVTNYFITS LACADLVMGL AVVPFGAAH ILMKMWTFGN FWCEFWTSID VLCVTASIET LCVIAVDRYF AITSPFKYQS LLTKNKARVI ILMVWIVSGL T SFLPIQMH ...String: MKTIIALSYI FCLVFADYKD DDDKDEVWVV GMGIVMSLIV LAIVFGNVLV ITAIAKFERL QTVTNYFITS LACADLVMGL AVVPFGAAH ILMKMWTFGN FWCEFWTSID VLCVTASIET LCVIAVDRYF AITSPFKYQS LLTKNKARVI ILMVWIVSGL T SFLPIQMH WYRATHQEAI NCYAEETCCD FFTNQAYAIA SSIVSFYVPL VIMVFVYSRV FQEAKRQLAD LEDNWETLND NL KVIEKAD NAAQVKDALT KMRAAALDAQ KASGSGSPEM KDFRHGFDIL VGQIDDALKL ANEGKVKEAQ AAAEQLKTTR NAY IQKYLK FCLKEHKALK TLGIIMGTFT LCWLPFFIVN IVHVIQDNLI RKEVYILLNW IGYVNSGFNP LIYSRSPDFR IAFQ ELLKI AALKEKIAAL KEKIAALKEA EEKRASRLEE ELRRRLTEGS HHHHHHHH UniProtKB: Beta-2 adrenergic receptor, Beta-2 adrenergic receptor |
-Macromolecule #2: (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol
| Macromolecule | Name: (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol type: ligand / ID: 2 / Number of copies: 1 / Formula: JTZ |
|---|---|
| Molecular weight | Theoretical: 249.349 Da |
| Chemical component information | 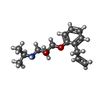 ChemComp-JTZ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: NICKEL/TITANIUM / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: NITROGEN / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)