-Search query
-Search result
Showing 1 - 50 of 374 items for (author: gad & h)

EMDB-16543: 
Structure of human mitochondrial RNase P in complex with mitochondrial pre-tRNA-His(5,Ser)

EMDB-16544: 
Structure of human mitochondrial RNase Z in complex with mitochondrial pre-tRNA-His(0,Ser)

EMDB-16545: 
Structure of human mitochondrial CCA-adding enzyme in complex with mitochondrial pre-tRNA-Ile

EMDB-16547: 
Structure of human mitochondrial MRPP1-MRPP2 in complex with mitochondrial pre-tRNA-Ile

EMDB-16667: 
Consensus Refinement - Map 1

EMDB-16668: 
Local refinement - Map 2

EMDB-16669: 
Cryo-EM structure of human mitochondrial RNase P in complex with mitochondrial pre-tRNA-His(5,Ser)

EMDB-16673: 
Consensus Refinement - Map 1

EMDB-16674: 
Local refinement - Map 2 (mask 1)

EMDB-16675: 
Local refinement - map 3 (Mask 2)

EMDB-16690: 
Consensus Refinement - Map 1

EMDB-16691: 
Local refinement - Map 2 (Mask 1)

EMDB-16692: 
Global Refinement - Map 1

PDB-8cbk: 
Structure of human mitochondrial RNase P in complex with mitochondrial pre-tRNA-His(5,Ser)

PDB-8cbl: 
Structure of human mitochondrial RNase Z in complex with mitochondrial pre-tRNA-His(0,Ser)

PDB-8cbm: 
Structure of human mitochondrial CCA-adding enzyme in complex with mitochondrial pre-tRNA-Ile

PDB-8cbo: 
Structure of human mitochondrial MRPP1-MRPP2 in complex with mitochondrial pre-tRNA-Ile

EMDB-18267: 
Structure of the human 20S U5 snRNP core

EMDB-19041: 
Structure of the human 20S U5 snRNP

PDB-8q91: 
Structure of the human 20S U5 snRNP core

PDB-8rc0: 
Structure of the human 20S U5 snRNP

EMDB-16005: 
GABA-A receptor a5 homomer - a5V3 - APO

EMDB-16050: 
GABA-A receptor a5 homomer - a5V3 - Basmisanil - HR

EMDB-16051: 
GABA-A receptor a5 homomer - a5V3 - RO154513

EMDB-16055: 
GABA-A receptor a5 homomer - a5V3 - RO5211223

EMDB-16058: 
GABA-A receptor a5 homomer - a5V3 - Diazepam

EMDB-16060: 
GABA-A receptor a5 homomer - a5V3 - DMCM

EMDB-16063: 
GABA-A receptor a5 homomer - a5V3 - L655708

EMDB-16066: 
GABA-A receptor a5 homomer - a5V3 - RO7172670

EMDB-16067: 
GABA-A receptor a5 homomer - a5V3 - RO7015738

EMDB-16068: 
GABA-A receptor a5 homomer - a5V3 - RO4938581

PDB-8bej: 
GABA-A receptor a5 homomer - a5V3 - APO

PDB-8bha: 
GABA-A receptor a5 homomer - a5V3 - Basmisanil - HR

PDB-8bhb: 
GABA-A receptor a5 homomer - a5V3 - RO154513

PDB-8bhi: 
GABA-A receptor a5 homomer - a5V3 - RO5211223

PDB-8bhk: 
GABA-A receptor a5 homomer - a5V3 - Diazepam

PDB-8bhm: 
GABA-A receptor a5 homomer - a5V3 - DMCM

PDB-8bho: 
GABA-A receptor a5 homomer - a5V3 - L655708

PDB-8bhq: 
GABA-A receptor a5 homomer - a5V3 - RO7172670

PDB-8bhr: 
GABA-A receptor a5 homomer - a5V3 - RO7015738

PDB-8bhs: 
GABA-A receptor a5 homomer - a5V3 - RO4938581

EMDB-18150: 
Cryo-EM map of the Candida albicans 80S ribosome in complex with cephaeline

EMDB-18151: 
Focused map on the body of the SSU of the Candida albicans 80S ribosome in complex with cephaeline

EMDB-18155: 
The cryo-EM map of the C. albicans ribosome focused on the head of the SSU

EMDB-18156: 
Combined map of Candida albicans 80S ribosome in complex with cephaeline
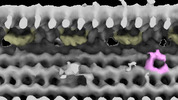
EMDB-28802: 
Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei FAP106 knockdown mutant
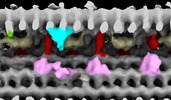
EMDB-28803: 
Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei Wildtype

EMDB-28804: 
Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei MC8 Knockdown mutant
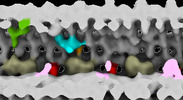
EMDB-28805: 
Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei Wildtype Control 2

EMDB-16145: 
Cryo-EM structure of NHEJ supercomplex(trimer)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
