-検索条件
-検索結果
検索 (著者・登録者: dietrich & l)の結果53件中、1から50件目までを表示しています

EMDB-50000: 
In situ structure of mitochondrial ATPsynthase in whole Polytomella cells

EMDB-50001: 
Structure of the peripheral stalk of the Polytomella ATPsynthase dimer in whole cells

EMDB-19999: 
In situ structure of the peripheral stalk of the mitochondrial ATPsynthase in whole Polytomella cells

PDB-9evd: 
In situ structure of the peripheral stalk of the mitochondrial ATPsynthase in whole Polytomella cells

EMDB-42434: 
Cryo-EM of (L, L)-2NapFF micelle

EMDB-42436: 
Cryo-EM of (L,D)-2NapFF micelle

EMDB-16451: 
Subtomogram average of the T. kivui 70S ribosome in situ

EMDB-14169: 
Cryo-EM structure of Hydrogen-dependent CO2 reductase.

EMDB-15053: 
In situ structure of HDCR filaments

EMDB-15054: 
In situ structure of the T. kivui ribosome

EMDB-15055: 
In situ cryo-electron tomogram of a T. kivui cell 1

EMDB-15056: 
In situ cryo-electron tomogram of a T. kivui cell 2

PDB-7qv7: 
Cryo-EM structure of Hydrogen-dependent CO2 reductase.

EMDB-13246: 
cryoFIB milling/cryoET of amoeba infected with Legionella pneumophila

EMDB-13247: 
cryoFIB milling/cryoET of amoeba infected with Legionella pneumophila

EMDB-13248: 
cryoFIB milling/cryoET of amoeba infected with Legionella pneumophila

EMDB-13249: 
cryoFIB milling/cryoET of amoeba infected with Legionella pneumophila

EMDB-24642: 
SARS-CoV-2 Spike bound to Fab PDI 210

EMDB-24643: 
SARS-CoV-2 Spike bound to Fab PDI 96

EMDB-24644: 
SARS-CoV-2 Spike bound to Fab PDI 215

EMDB-24645: 
SARS-CoV-2 Spike bound to Fab WCSL 119

EMDB-24646: 
SARS-CoV-2 Spike bound to Fab WCSL 129

EMDB-24647: 
SARS-CoV-2 Spike bound to Fab PDI 93

EMDB-24648: 
SARS-CoV-2 Spike bound to Fab PDI 222

EMDB-24649: 
SARS-CoV-2 receptor binding domain bound to Fab PDI 222

PDB-7rr0: 
SARS-CoV-2 receptor binding domain bound to Fab PDI 222

EMDB-11860: 
Subtomogram average structure of anammoxosomal nitrite oxidoreductase

EMDB-11861: 
Helical reconstruction of nitrite oxidoreductase from anammox bacteria

EMDB-23566: 
The SARS-CoV-2 spike protein receptor binding domain bound to neutralizing nanobodies WNb 2 and WNb 10

EMDB-23567: 
Cryo-EM map of SARS-CoV-2 Spike protein bound to neutralising nanobodies WNb2 and WNb10 (C3 symmetry)

EMDB-23568: 
Cryo-EM map of SARS-CoV-2 Spike protein in complex with neutralising nanobodies WNb2 and WNb10 (C1 symmetry)

PDB-7lx5: 
The SARS-CoV-2 spike protein receptor binding domain bound to neutralizing nanobodies WNb 2 and WNb 10

EMDB-11997: 
Structure of a nanoparticle for a COVID-19 vaccine candidate

PDB-7b3y: 
Structure of a nanoparticle for a COVID-19 vaccine candidate

EMDB-10840: 
Structure of RyR1 in apo/closed state in native membrane

EMDB-20348: 
EM structure of human tumor suppressor BRCA2 protein bound to human DSS1 protein and ssDNA without crosslinking

EMDB-21998: 
EM structure of human tumor suppressor bound to human DSS1 protein and ssDNA with crosslinking

EMDB-10834: 
Tobacco mosaic virus (TMV) structure determined by a hybrid STA-SPA workflow

EMDB-4584: 
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
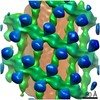
EMDB-10062: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
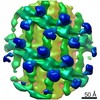
EMDB-10063: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state

EMDB-10064: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes

EMDB-10065: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state

PDB-6rzt: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes

PDB-6rzu: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state

PDB-6rzv: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes

PDB-6rzw: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state

EMDB-2657: 
TEM of uncleaved soluble HIV envelope proteins

EMDB-2659: 
TEM of uncleaved soluble HIV envelope proteins
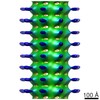
EMDB-2094: 
Central hub structure of Trichonympha cartwheel
ページ:
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア EMN検索について
EMN検索について



 wwPDBはEMDBデータモデルのバージョン3へ移行します
wwPDBはEMDBデータモデルのバージョン3へ移行します
