[English] 日本語
 Yorodumi
Yorodumi- EMDB-10834: Tobacco mosaic virus (TMV) structure determined by a hybrid STA-S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10834 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tobacco mosaic virus (TMV) structure determined by a hybrid STA-SPA workflow | |||||||||
 Map data Map data | refine map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Tobacco mosaic virus-like, coat protein / Tobacco mosaic virus-like, coat protein superfamily / Virus coat protein (TMV like) / helical viral capsid / structural molecule activity / identical protein binding / Capsid protein / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   Tobacco mosaic virus Tobacco mosaic virus | |||||||||
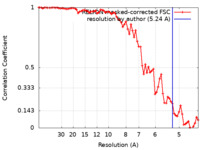
| Method | subtomogram averaging / cryo EM / Resolution: 5.24 Å | |||||||||
 Authors Authors | Sanchez RM / Zhang Y / Chen W / Dietrich L / Kudryashev M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Subnanometer-resolution structure determination in situ by hybrid subtomogram averaging - single particle cryo-EM. Authors: Ricardo M Sanchez / Yingyi Zhang / Wenbo Chen / Lea Dietrich / Mikhail Kudryashev /  Abstract: Cryo-electron tomography combined with subtomogram averaging (StA) has yielded high-resolution structures of macromolecules in their native context. However, high-resolution StA is not commonplace ...Cryo-electron tomography combined with subtomogram averaging (StA) has yielded high-resolution structures of macromolecules in their native context. However, high-resolution StA is not commonplace due to beam-induced sample drift, images with poor signal-to-noise ratios (SNR), challenges in CTF correction, and limited particle number. Here we address these issues by collecting tilt series with a higher electron dose at the zero-degree tilt. Particles of interest are then located within reconstructed tomograms, processed by conventional StA, and then re-extracted from the high-dose images in 2D. Single particle analysis tools are then applied to refine the 2D particle alignment and generate a reconstruction. Use of our hybrid StA (hStA) workflow improved the resolution for tobacco mosaic virus from 7.2 to 4.4 Å and for the ion channel RyR1 in crowded native membranes from 12.9 to 9.1 Å. These resolution gains make hStA a promising approach for other StA projects aimed at achieving subnanometer resolution. #2:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Subnanometer-resolution structure determination in situ by a hybrid subtomogram averaging - single particle cryoEM - workflow Authors: Sanchez RM / Zhang Y / Chen W / Dietrich L / Kudryashev M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10834.map.gz emd_10834.map.gz | 22.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10834-v30.xml emd-10834-v30.xml emd-10834.xml emd-10834.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10834_fsc.xml emd_10834_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10834.png emd_10834.png | 35.3 KB | ||
| Masks |  emd_10834_msk_1.map emd_10834_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Others |  emd_10834_half_map_1.map.gz emd_10834_half_map_1.map.gz emd_10834_half_map_2.map.gz emd_10834_half_map_2.map.gz | 23.1 MB 23.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10834 http://ftp.pdbj.org/pub/emdb/structures/EMD-10834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10834 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10393 (Title: Subnanometer-resolution structure determination in situ by a hybrid subtomogram averaging - single particle cryoEM - workflow - on TMV EMPIAR-10393 (Title: Subnanometer-resolution structure determination in situ by a hybrid subtomogram averaging - single particle cryoEM - workflow - on TMVData size: 37.5 Data #1: TMV-hybrid tomography- tiltstacks- aligned and CTF corrected [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10834.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10834.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refine map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
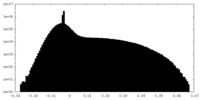
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10834_msk_1.map emd_10834_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
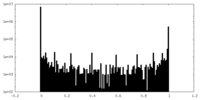
| Density Histograms |
-Half map: half map 1
| File | emd_10834_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
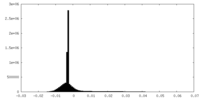
| Density Histograms |
-Half map: half map 2
| File | emd_10834_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
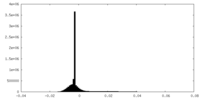
| Density Histograms |
- Sample components
Sample components
-Entire : Tobacco mosaic virus
| Entire | Name:   Tobacco mosaic virus Tobacco mosaic virus |
|---|---|
| Components |
|
-Supramolecule #1: Tobacco mosaic virus
| Supramolecule | Name: Tobacco mosaic virus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 12242 / Sci species name: Tobacco mosaic virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: Capsid protein / Diameter: 180.0 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 33 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 2.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -0.0045 µm / Nominal defocus min: -0.0035 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)