[English] 日本語
 Yorodumi
Yorodumi- PDB-6rzt: Structure of s-Mgm1 decorating the outer surface of tubulated lip... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6rzt | ||||||
|---|---|---|---|---|---|---|---|
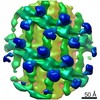
| Title | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | ||||||
 Components Components | Putative mitochondrial dynamin protein | ||||||
 Keywords Keywords | CONTRACTILE PROTEIN / mitochondrial protein / membrane remodelling / GTPase / mitochondrial dynamics / subtomogram averaging | ||||||
| Function / homology |  Function and homology information Function and homology informationmembrane bending activity / GTPase-dependent fusogenic activity / dynamin GTPase / receptor internalization / mitochondrial intermembrane space / microtubule binding / microtubule / mitochondrial inner membrane / GTPase activity / lipid binding ...membrane bending activity / GTPase-dependent fusogenic activity / dynamin GTPase / receptor internalization / mitochondrial intermembrane space / microtubule binding / microtubule / mitochondrial inner membrane / GTPase activity / lipid binding / GTP binding / metal ion binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | ||||||
| Method | ELECTRON MICROSCOPY / subtomogram averaging / cryo EM / Resolution: 14.7 Å | ||||||
 Authors Authors | Faelber, K. / Dietrich, L. / Noel, J.K. / Sanchez, R. / Kudryashev, M. / Kuehlbrandt, W. / Daumke, O. | ||||||
| Funding support |  Germany, 1items Germany, 1items
| ||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Authors: Katja Faelber / Lea Dietrich / Jeffrey K Noel / Florian Wollweber / Anna-Katharina Pfitzner / Alexander Mühleip / Ricardo Sánchez / Misha Kudryashev / Nicolas Chiaruttini / Hauke Lilie / ...Authors: Katja Faelber / Lea Dietrich / Jeffrey K Noel / Florian Wollweber / Anna-Katharina Pfitzner / Alexander Mühleip / Ricardo Sánchez / Misha Kudryashev / Nicolas Chiaruttini / Hauke Lilie / Jeanette Schlegel / Eva Rosenbaum / Manuel Hessenberger / Claudia Matthaeus / Séverine Kunz / Alexander von der Malsburg / Frank Noé / Aurélien Roux / Martin van der Laan / Werner Kühlbrandt / Oliver Daumke /   Abstract: Balanced fusion and fission are key for the proper function and physiology of mitochondria. Remodelling of the mitochondrial inner membrane is mediated by the dynamin-like protein mitochondrial ...Balanced fusion and fission are key for the proper function and physiology of mitochondria. Remodelling of the mitochondrial inner membrane is mediated by the dynamin-like protein mitochondrial genome maintenance 1 (Mgm1) in fungi or the related protein optic atrophy 1 (OPA1) in animals. Mgm1 is required for the preservation of mitochondrial DNA in yeast, whereas mutations in the OPA1 gene in humans are a common cause of autosomal dominant optic atrophy-a genetic disorder that affects the optic nerve. Mgm1 and OPA1 are present in mitochondria as a membrane-integral long form and a short form that is soluble in the intermembrane space. Yeast strains that express temperature-sensitive mutants of Mgm1 or mammalian cells that lack OPA1 display fragmented mitochondria, which suggests that Mgm1 and OPA1 have an important role in inner-membrane fusion. Consistently, only the mitochondrial outer membrane-not the inner membrane-fuses in the absence of functional Mgm1. Mgm1 and OPA1 have also been shown to maintain proper cristae architecture; for example, OPA1 prevents the release of pro-apoptotic factors by tightening crista junctions. Finally, the short form of OPA1 localizes to mitochondrial constriction sites, where it presumably promotes mitochondrial fission. How Mgm1 and OPA1 perform their diverse functions in membrane fusion, scission and cristae organization is at present unknown. Here we present crystal and electron cryo-tomography structures of Mgm1 from Chaetomium thermophilum. Mgm1 consists of a GTPase (G) domain, a bundle signalling element domain, a stalk, and a paddle domain that contains a membrane-binding site. Biochemical and cell-based experiments demonstrate that the Mgm1 stalk mediates the assembly of bent tetramers into helical filaments. Electron cryo-tomography studies of Mgm1-decorated lipid tubes and fluorescence microscopy experiments on reconstituted membrane tubes indicate how the tetramers assemble on positively or negatively curved membranes. Our findings convey how Mgm1 and OPA1 filaments dynamically remodel the mitochondrial inner membrane. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6rzt.cif.gz 6rzt.cif.gz | 1.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6rzt.ent.gz pdb6rzt.ent.gz | 910.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6rzt.json.gz 6rzt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rz/6rzt https://data.pdbj.org/pub/pdb/validation_reports/rz/6rzt ftp://data.pdbj.org/pub/pdb/validation_reports/rz/6rzt ftp://data.pdbj.org/pub/pdb/validation_reports/rz/6rzt | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 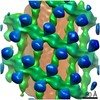 10062MC  4584C  6ql4C  6rzuC  6rzvC  6rzwC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 77360.172 Da / Num. of mol.: 12 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Chaetomium thermophilum (fungus) / Gene: CTHT_0065850 / Production host: Chaetomium thermophilum (fungus) / Gene: CTHT_0065850 / Production host:  Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: subtomogram averaging |
- Sample preparation
Sample preparation
| Component | Name: Mgm1 / Type: COMPLEX / Details: short isoform with C- and N-terminal truncations / Entity ID: all / Source: RECOMBINANT |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.4 / Details: 20 mM HEPES, 200 mM NaCl, residual MgCl2, 9mM KCl. |
| Specimen | Conc.: 5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: Sample was prepared in the described buffer. Just before freezing 6 nm colloidal gold fiducial marker were added to the sample in a 1:1 ratio. |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 70 % / Chamber temperature: 277.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 53000 X / Nominal defocus max: 4000 nm / Nominal defocus min: 2000 nm / Cs: 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: BASIC |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 2 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
| EM imaging optics | Energyfilter slit width: 20 eV |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: CTF determination was done by Gctf and correction was performed by ctfphaseflip from IMOD Type: PHASE FLIPPING ONLY | ||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 14.7 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 11474 Details: Half sets were generated not even-odd but as upper and lower-halves of particles in given tomogram Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| EM volume selection | Method: Geometry-assisted particle picking form tube surfaces Details: with the use of Dynamo Catalogue system / Num. of tomograms: 15 / Num. of volumes extracted: 12440 Reference model: global average of the particles rotated to the known initial orientations |
 Movie
Movie Controller
Controller






 PDBj
PDBj