[English] 日本語
 Yorodumi
Yorodumi- EMDB-21998: EM structure of human tumor suppressor bound to human DSS1 protei... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21998 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
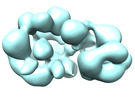
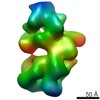
| Title | EM structure of human tumor suppressor bound to human DSS1 protein and ssDNA with crosslinking | ||||||||||||
 Map data Map data | Human BRCA2-DSS1-ssDNA complex with crosslinking | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / negative staining / Resolution: 14.6 Å | ||||||||||||
 Authors Authors | Le HP / Ma X / Vaquero J / Brinkmeyer M / Guo F / Heyer W-D / Liu J | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2020 Journal: Nucleic Acids Res / Year: 2020Title: DSS1 and ssDNA regulate oligomerization of BRCA2. Authors: Hang Phuong Le / Xiaoyan Ma / Jorge Vaquero / Megan Brinkmeyer / Fei Guo / Wolf-Dietrich Heyer / Jie Liu /  Abstract: The tumor suppressor BRCA2 plays a key role in initiating homologous recombination by facilitating RAD51 filament formation on single-stranded DNA. The small acidic protein DSS1 is a crucial partner ...The tumor suppressor BRCA2 plays a key role in initiating homologous recombination by facilitating RAD51 filament formation on single-stranded DNA. The small acidic protein DSS1 is a crucial partner to BRCA2 in this process. In vitro and in cells (1,2), BRCA2 associates into oligomeric complexes besides also existing as monomers. A dimeric structure was further characterized by electron microscopic analysis (3), but the functional significance of the different BRCA2 assemblies remains to be determined. Here, we used biochemistry and electron microscopic imaging to demonstrate that the multimerization of BRCA2 is counteracted by DSS1 and ssDNA. When validating the findings, we identified three self-interacting regions and two types of self-association, the N-to-C terminal and the N-to-N terminal interactions. The N-to-C terminal self-interaction of BRCA2 is sensitive to DSS1 and ssDNA. The N-to-N terminal self-interaction is modulated by ssDNA. Our results define a novel role of DSS1 to regulate BRCA2 in an RPA-independent fashion. Since DSS1 is required for BRCA2 function in recombination, we speculate that the monomeric and oligomeric forms of BRCA2 might be active for different cellular events in recombinational DNA repair and replication fork stabilization. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21998.map.gz emd_21998.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21998-v30.xml emd-21998-v30.xml emd-21998.xml emd-21998.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21998_fsc.xml emd_21998_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_21998.png emd_21998.png | 104.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21998 http://ftp.pdbj.org/pub/emdb/structures/EMD-21998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21998 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21998.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21998.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human BRCA2-DSS1-ssDNA complex with crosslinking | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.48 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BRCA2-DSS1-ssDNA
| Entire | Name: BRCA2-DSS1-ssDNA |
|---|---|
| Components |
|
-Supramolecule #1: BRCA2-DSS1-ssDNA
| Supramolecule | Name: BRCA2-DSS1-ssDNA / type: complex / ID: 1 / Parent: 0 Details: Full-length human tumor suppressor BRCA2 protein in complex with DSS1 and dT100 oligo with crosslinking |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
| Molecular weight | Experimental: 474 kDa/nm |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Staining | Type: NEGATIVE / Material: 2% uranyl acetate Details: BRCA2 complex was applied to glow-discharged continuous carbon copper grids and incubated for 2 min. Grids were washed briefly with 5 mM Mg(Acet)2 for three times to remove buffer, sucrose, ...Details: BRCA2 complex was applied to glow-discharged continuous carbon copper grids and incubated for 2 min. Grids were washed briefly with 5 mM Mg(Acet)2 for three times to remove buffer, sucrose, and glycerol. Samples were then stained with 2% uranyl acetate, blotted with filter paper, and air dried. |
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100F |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Digitization - Dimensions - Width: 5120 pixel / Digitization - Dimensions - Height: 3840 pixel / Digitization - Sampling interval: 6.4 µm / Number grids imaged: 1 / Number real images: 1359 / Average exposure time: 1.0 sec. / Average electron dose: 37.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder model: JEOL |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















