+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of (L,D)-2NapFF micelle | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dipeptides / micelle / stereoisomers / PROTEIN FIBRIL | |||||||||
| Biological species | synthetic construct (others) | |||||||||
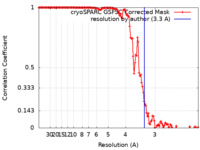
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Sonani RR / Adams DJ / Egelman EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep Phys Sci / Year: 2024 Journal: Cell Rep Phys Sci / Year: 2024Title: Atomic structures of naphthalene dipeptide micelles unravel mechanisms of assembly and gelation. Authors: Ravi R Sonani / Simona Bianco / Bart Dietrich / James Doutch / Emily R Draper / Dave J Adams / Edward H Egelman /   Abstract: Peptide-based biopolymers have gained increasing attention due to their versatile applications. A naphthalene dipeptide (2NapFF) can form chirality-dependent tubular micelles, leading to ...Peptide-based biopolymers have gained increasing attention due to their versatile applications. A naphthalene dipeptide (2NapFF) can form chirality-dependent tubular micelles, leading to supramolecular gels. The precise molecular arrangement within these micelles and the mechanism governing gelation have remained enigmatic. We determined, at near-atomic resolution, cryoelectron microscopy structures of the 2NapFF micelles LL-tube and LD-tube, generated by the stereoisomers (l,l)-2NapFF and (l,d)-2NapFF, respectively. The structures reveal that the fundamental packing of dipeptides is driven by the systematic π-π stacking of aromatic rings and that same-charge repulsion between the carbonyl groups is responsible for the stiffness of both tubes. The structural analysis elucidates how a single residue's altered chirality gives rise to markedly distinct tubular structures and sheds light on the mechanisms underlying the pH-dependent gelation of LL- and LD-tubes. The understanding of dipeptide packing and gelation mechanisms provides insights for the rational design of 2NapFF derivatives, enabling the modulation of micellar dimensions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42436.map.gz emd_42436.map.gz | 189 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42436-v30.xml emd-42436-v30.xml emd-42436.xml emd-42436.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42436_fsc.xml emd_42436_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42436.png emd_42436.png | 236.9 KB | ||
| Filedesc metadata |  emd-42436.cif.gz emd-42436.cif.gz | 4 KB | ||
| Others |  emd_42436_half_map_1.map.gz emd_42436_half_map_1.map.gz emd_42436_half_map_2.map.gz emd_42436_half_map_2.map.gz | 199.2 MB 199.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42436 http://ftp.pdbj.org/pub/emdb/structures/EMD-42436 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42436 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42436 | HTTPS FTP |
-Validation report
| Summary document |  emd_42436_validation.pdf.gz emd_42436_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42436_full_validation.pdf.gz emd_42436_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_42436_validation.xml.gz emd_42436_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_42436_validation.cif.gz emd_42436_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42436 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42436 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42436 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42436 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42436.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42436.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_42436_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_42436_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : (L, L)-2NapFF micelle
| Entire | Name: (L, L)-2NapFF micelle |
|---|---|
| Components |
|
-Supramolecule #1: (L, L)-2NapFF micelle
| Supramolecule | Name: (L, L)-2NapFF micelle / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) / Synthetically produced: Yes |
-Macromolecule #1: (L,D)-2NapFF dipeptide
| Macromolecule | Name: (L,D)-2NapFF dipeptide / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: (NwF)(DPH) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 11 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)