-Search query
-Search result
Showing all 50 items for (author: bhatt & pr)

EMDB-29723: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate, 30S focused map

EMDB-29724: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate consensus, 30S focused

EMDB-29833: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 30S focused

EMDB-29834: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate, 50S focused from 30S subtracted

EMDB-29842: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate, 50S focused and 30S subtracted

EMDB-29681: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms

EMDB-29687: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate

EMDB-29688: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate

EMDB-29689: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate

EMDB-29844: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate, 50S focused and 30S subtracted

PDB-8g2u: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms

PDB-8g31: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate

PDB-8g34: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate

PDB-8g38: 
Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate

EMDB-15793: 
Translating 70S ribosome in the unrotated state (P and E, tRNAs)

PDB-8b0x: 
Translating 70S ribosome in the unrotated state (P and E, tRNAs)

EMDB-15794: 
Translating 70S ribosome in the rotated state (A/A and P/E tRNAs)

EMDB-15795: 
Translating 70S ribosome in the rotated state (A/P and P/E tRNAs)

EMDB-15796: 
Translating 70S ribosome in the unrotated state (A and P tRNAs)

EMDB-15797: 
Translating 70S ribosome in the unrotated state (A,P and E tRNAs)
EMDB-26467: 
SARS-CoV-2 6P Mut7 in complex with Fab THSC20.HVTR04 (1 RBD up and 1 RDB down)

EMDB-26470: 
SARS-CoV-2 6P Mut7 in complex with Fab THSC20.HVTR26 (1 RBD up, 1 RBD down)

EMDB-26472: 
SARS-CoV-2 6P Mut7 in complex with Fab THSC20.HVTR26 (1 RBD up)

EMDB-26473: 
SARS-CoV-2 6P Mut7 in complex with Fab THSC20.HVTR26 (3 RBD down)

EMDB-23280: 
Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand

EMDB-23281: 
Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist

PDB-7ld3: 
Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand

PDB-7ld4: 
Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist

EMDB-12756: 
Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution)

EMDB-12757: 
Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot)

EMDB-12758: 
Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site

EMDB-12759: 
Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot

EMDB-12760: 
Disome of rabbit ribosomes with the leading one stalled by the SARS-CoV-2 pseudoknot

PDB-7o7y: 
Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution)

PDB-7o7z: 
Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot)

PDB-7o80: 
Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site

PDB-7o81: 
Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot
EMDB-22496: 
Base-directed response of rabbit #1 week 6 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP

EMDB-22498: 
N611-site directed response of rabbit #1 week 6 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP

EMDB-22499: 
Base-directed response of rabbit #1 week 12 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
EMDB-22500: 
N611-site directed response of rabbit #1 week 12 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
EMDB-22501: 
C3/V5-directed response of rabbit #1 week 12 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
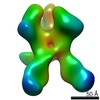
EMDB-22502: 
Base-directed response of rabbit #1 week 22 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
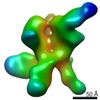
EMDB-22503: 
N611-site, base, and V1V3 directed response of rabbit #1 week 22 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
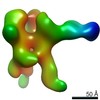
EMDB-22504: 
C3/V5 and base directed response of rabbit #1 week 22 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP

EMDB-22505: 
V1/V3 and base directed response of rabbit #1 week 22 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
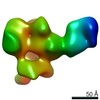
EMDB-23159: 
C3/V5-directed response of human donor G37080 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP

EMDB-0466: 
HIV-1 Env State 2A

PDB-3iyk: 
Bluetongue virus structure reveals a sialic acid binding domain, amphipathic helices and a central coiled coil in the outer capsid proteins

EMDB-5147: 
Bluetongue virus structure reveals a sialic acid binding domain, amphipathic helices and a central coiled coil in the outer capsid proteins
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
