[English] 日本語
 Yorodumi
Yorodumi- EMDB-23280: Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23280 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand | |||||||||
 Map data Map data | Final masked map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | signaling protein / membrane protein / active-state G protein-coupled receptor / adenosine A1 receptor / PAM / allosteric modulator | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of nucleoside transport / negative regulation of neurotrophin production / negative regulation of circadian sleep/wake cycle, non-REM sleep / regulation of glomerular filtration / purine nucleoside binding / G protein-coupled purinergic nucleotide receptor signaling pathway / negative regulation of mucus secretion / positive regulation of peptide secretion / negative regulation of glutamate secretion / negative regulation of synaptic transmission, GABAergic ...positive regulation of nucleoside transport / negative regulation of neurotrophin production / negative regulation of circadian sleep/wake cycle, non-REM sleep / regulation of glomerular filtration / purine nucleoside binding / G protein-coupled purinergic nucleotide receptor signaling pathway / negative regulation of mucus secretion / positive regulation of peptide secretion / negative regulation of glutamate secretion / negative regulation of synaptic transmission, GABAergic / negative regulation of long-term synaptic depression / positive regulation of dephosphorylation / positive regulation of lipid catabolic process / negative regulation of hormone secretion / negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway / negative regulation of leukocyte migration / mucus secretion / Muscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / Adenosine P1 receptors / regulation of sensory perception of pain / heterotrimeric G-protein binding / regulation of respiratory gaseous exchange by nervous system process / G protein-coupled adenosine receptor activity / response to purine-containing compound / regulation of presynaptic cytosolic calcium ion concentration / negative regulation of calcium ion-dependent exocytosis / G protein-coupled adenosine receptor signaling pathway / negative regulation of adenylate cyclase activity / positive regulation of potassium ion transport / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / negative regulation of systemic arterial blood pressure / negative regulation of synaptic transmission / positive regulation of neural precursor cell proliferation / regulation of cardiac muscle cell contraction / long-term synaptic depression / presynaptic active zone / positive regulation of urine volume / negative regulation of synaptic transmission, glutamatergic / protein targeting to membrane / triglyceride homeostasis / leukocyte migration / gamma-aminobutyric acid signaling pathway / regulation of locomotion / temperature homeostasis / detection of temperature stimulus involved in sensory perception of pain / negative regulation of acute inflammatory response / positive regulation of systemic arterial blood pressure / regulation of calcium ion transport / negative regulation of long-term synaptic potentiation / asymmetric synapse / negative regulation of apoptotic signaling pathway / axolemma / fatty acid homeostasis / phagocytosis / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / negative regulation of lipid catabolic process / neuronal dense core vesicle / lipid catabolic process / positive regulation of vascular associated smooth muscle cell proliferation / positive regulation of superoxide anion generation / heat shock protein binding / Adenylate cyclase inhibitory pathway / response to nutrient / calyx of Held / hippocampal mossy fiber to CA3 synapse / excitatory postsynaptic potential / apoptotic signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / vasodilation / cognition / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / terminal bouton / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
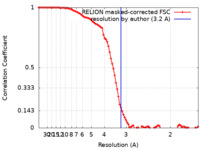
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Draper-Joyce CJ / Danev R | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Positive allosteric mechanisms of adenosine A receptor-mediated analgesia. Authors: Christopher J Draper-Joyce / Rebecca Bhola / Jinan Wang / Apurba Bhattarai / Anh T N Nguyen / India Cowie-Kent / Kelly O'Sullivan / Ling Yeong Chia / Hariprasad Venugopal / Celine Valant / ...Authors: Christopher J Draper-Joyce / Rebecca Bhola / Jinan Wang / Apurba Bhattarai / Anh T N Nguyen / India Cowie-Kent / Kelly O'Sullivan / Ling Yeong Chia / Hariprasad Venugopal / Celine Valant / David M Thal / Denise Wootten / Nicolas Panel / Jens Carlsson / Macdonald J Christie / Paul J White / Peter Scammells / Lauren T May / Patrick M Sexton / Radostin Danev / Yinglong Miao / Alisa Glukhova / Wendy L Imlach / Arthur Christopoulos /     Abstract: The adenosine A receptor (AR) is a promising therapeutic target for non-opioid analgesic agents to treat neuropathic pain. However, development of analgesic orthosteric AR agonists has failed because ...The adenosine A receptor (AR) is a promising therapeutic target for non-opioid analgesic agents to treat neuropathic pain. However, development of analgesic orthosteric AR agonists has failed because of a lack of sufficient on-target selectivity as well as off-tissue adverse effects. Here we show that [2-amino-4-(3,5-bis(trifluoromethyl)phenyl)thiophen-3-yl)(4-chlorophenyl)methanone] (MIPS521), a positive allosteric modulator of the AR, exhibits analgesic efficacy in rats in vivo through modulation of the increased levels of endogenous adenosine that occur in the spinal cord of rats with neuropathic pain. We also report the structure of the AR co-bound to adenosine, MIPS521 and a G heterotrimer, revealing an extrahelical lipid-detergent-facing allosteric binding pocket that involves transmembrane helixes 1, 6 and 7. Molecular dynamics simulations and ligand kinetic binding experiments support a mechanism whereby MIPS521 stabilizes the adenosine-receptor-G protein complex. This study provides proof of concept for structure-based allosteric drug design of non-opioid analgesic agents that are specific to disease contexts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23280.map.gz emd_23280.map.gz | 6.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23280-v30.xml emd-23280-v30.xml emd-23280.xml emd-23280.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23280_fsc.xml emd_23280_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_23280.png emd_23280.png | 187.9 KB | ||
| Masks |  emd_23280_msk_1.map emd_23280_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23280.cif.gz emd-23280.cif.gz | 6.9 KB | ||
| Others |  emd_23280_additional_1.map.gz emd_23280_additional_1.map.gz emd_23280_half_map_1.map.gz emd_23280_half_map_1.map.gz emd_23280_half_map_2.map.gz emd_23280_half_map_2.map.gz | 49.2 MB 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23280 http://ftp.pdbj.org/pub/emdb/structures/EMD-23280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23280 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23280 | HTTPS FTP |
-Related structure data
| Related structure data |  7ld3MC  7ld4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23280.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23280.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final masked map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23280_msk_1.map emd_23280_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Final Refine3D map
| File | emd_23280_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final Refine3D map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Final Refine3D halfmap 1
| File | emd_23280_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final Refine3D halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Final Refine3D halfmap 1
| File | emd_23280_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final Refine3D halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human adenosine A1 receptor-Gi2-protein complex bound to its endo...
| Entire | Name: Human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist adenosine |
|---|---|
| Components |
|
-Supramolecule #1: Human adenosine A1 receptor-Gi2-protein complex bound to its endo...
| Supramolecule | Name: Human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist adenosine type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-2 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.502863 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTVSAEDK AAAERSKMID KNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEDG YSEEECRQYR AVVYSNTIQS IMAIVKAMG NLQIDFADPS RADDARQLFA LSCTAEEQGV LPDDLSGVIR RLWADHGVQA CFGRSREYQL NDSAAYYLND L ERIAQSDY ...String: MGCTVSAEDK AAAERSKMID KNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEDG YSEEECRQYR AVVYSNTIQS IMAIVKAMG NLQIDFADPS RADDARQLFA LSCTAEEQGV LPDDLSGVIR RLWADHGVQA CFGRSREYQL NDSAAYYLND L ERIAQSDY IPTQQDVLRT RVKTTGIVET HFTFKDLHFK MFDVGAQRSE RKKWIHCFEG VTAIIFCVAL SAYDLVLAED EE MNRMHAS MKLFDSICNN KWFTDTSIIL FLNKKDLFEE KITHSPLTIC FPEYTGANKY DEAASYIQSK FEDLNKRKDT KEI YTHFTC STDTKNVQFV FDAVTDVIIK NNLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-2 |
-Macromolecule #2: Chimera protein of Muscarinic acetylcholine receptor M4 and Adeno...
| Macromolecule | Name: Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.26102 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAMGANFT PVNGSSGNQS VRLVTSSSLE VLFQGPPPSI SAFQAAYIGI EVLIALVSVP GNVLVIWAV KVNQALRDAT FCFIVSLAVA DVAVGALVIP LAILINIGPQ TYFHTCLMVA CPVLILTQSS ILALLAIAVD R YLRVKIPL ...String: MKTIIALSYI FCLVFADYKD DDDAMGANFT PVNGSSGNQS VRLVTSSSLE VLFQGPPPSI SAFQAAYIGI EVLIALVSVP GNVLVIWAV KVNQALRDAT FCFIVSLAVA DVAVGALVIP LAILINIGPQ TYFHTCLMVA CPVLILTQSS ILALLAIAVD R YLRVKIPL RYKMVVTPRR AAVAIAGCWI LSFVVGLTPM FGWNNLSAVE RAWAANGSMG EPVIKCEFEK VISMEYMVYF NF FVWVLPP LLLMVLIYLE VFYLIRKQLN KKVSASSGDP QKYYGKELKI AKSLALILFL FALSWLPLHI LNCITLFCPS CHK PSILTY IAIFLTHGNS AMNPIVYAFR IQKFRVTFLK IWNDHFRCQP APPIDEDLPE ERPDDHHHHH H UniProtKB: Muscarinic acetylcholine receptor M4, Adenosine receptor A1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.534062 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: ADENOSINE
| Macromolecule | Name: ADENOSINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ADN |
|---|---|
| Molecular weight | Theoretical: 267.241 Da |
| Chemical component information |  ChemComp-ADN: |
-Macromolecule #6: {2-amino-4-[3,5-bis(trifluoromethyl)phenyl]thiophen-3-yl}(4-chlor...
| Macromolecule | Name: {2-amino-4-[3,5-bis(trifluoromethyl)phenyl]thiophen-3-yl}(4-chlorophenyl)methanone type: ligand / ID: 6 / Number of copies: 1 / Formula: XTD |
|---|---|
| Molecular weight | Theoretical: 449.797 Da |
| Chemical component information | 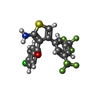 ChemComp-XTD: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average exposure time: 8.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)