-Search query
-Search result
Showing 1 - 50 of 6,724 items for (author: zhang & k)
EMDB-44255: 
Cryo-EM structure of the mouse TRPM8 channel in the ligand-free desensitized state
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY

EMDB-44256: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY

EMDB-44257: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMG2850
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
EMDB-44258: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMTB
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
EMDB-44259: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 and the cooling agonist C3
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
EMDB-44260: 
Cryo-EM structure of the avian great tit TRPM8 channel in complex with the antagonist TC-I 2014
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY

EMDB-44261: 
Cryo-EM structure of the mouse TRPM8 channel in complex with PI(4,5)P2 and Ca2+
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
EMDB-44262: 
Cryo-EM structure of the mouse TRPM8 channel in complex with Ca2+ in the absence of PI(4,5)P2
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
EMDB-60628: 
Carazolol-activated human beta3 adrenergic receptor
Method: single particle / : Zheng S, Zhang S, Dai S, Chen K, Gao K, Lin B, Liu X
EMDB-60629: 
Epinephrine-activated human beta3 adrenergic receptor
Method: single particle / : Zheng S, Zhang S, Dai S, Chen K, Gao K, Lin B, Liu X

EMDB-38695: 
Cryo-EM structure of ATP-DNA-MuB filaments
Method: helical / : Zhao X, Zhang K, Li S

EMDB-38696: 
CryoEM structure of ADP-DNA-MuB conformation1
Method: single particle / : Zhao X, Zhang K, Li S

EMDB-38697: 
CryoEM structure of ADP-DNA-MuB conformation2
Method: single particle / : Zhao X, Zhang K, Li S

EMDB-38698: 
CryoEM structure of ADP-DNA-MuB conformation3
Method: single particle / : Zhao X, Zhang K, Li S

EMDB-38699: 
CryoEM structure of ADP-DNA-MuB conformation4
Method: single particle / : Zhang X, Zhang K, Li S

PDB-8xvc: 
CryoEM structure of ADP-DNA-MuB conformation1
Method: single particle / : Zhao X, Zhang K, Li S

PDB-8xvd: 
CryoEM structure of ADP-DNA-MuB conformation2
Method: single particle / : Zhao X, Zhang K, Li S

EMDB-41770: 
Apo form of human ATE1
Method: single particle / : Huang W, Zhang Y, Taylor DJ

EMDB-42071: 
human ATE1 in complex with Arg-tRNA and a peptide substrate
Method: single particle / : Huang W, Zhang Y, Taylor DJ

PDB-8uau: 
human ATE1 in complex with Arg-tRNA and a peptide substrate
Method: single particle / : Huang W, Zhang Y, Taylor DJ

EMDB-37842: 
Cryo-EM structure of noradrenaline transporter in apo state
Method: single particle / : Zhao Y, Hu T, Yu Z
EMDB-37843: 
Cryo-EM structure of noradrenaline transporter in complex with noradrenaline
Method: single particle / : Zhao Y, Hu T, Yu Z
EMDB-37844: 
Cryo-EM structure of noradrenaline transporter in complex with a x-MrlA analogue
Method: single particle / : Zhao Y, Hu T, Yu Z
EMDB-37845: 
Cryo-EM structure of noradrenaline transporter in complex with bupropion
Method: single particle / : Zhao Y, Hu T, Yu Z
EMDB-37846: 
Cryo-EM structure of noradrenaline transporter in complex with ziprasidone
Method: single particle / : Zhao Y, Hu T, Yu Z

PDB-8wtu: 
Cryo-EM structure of noradrenaline transporter in apo state
Method: single particle / : Zhao Y, Hu T, Yu Z

PDB-8wtv: 
Cryo-EM structure of noradrenaline transporter in complex with noradrenaline
Method: single particle / : Zhao Y, Hu T, Yu Z

PDB-8wtw: 
Cryo-EM structure of noradrenaline transporter in complex with a x-MrlA analogue
Method: single particle / : Zhao Y, Hu T, Yu Z

PDB-8wtx: 
Cryo-EM structure of noradrenaline transporter in complex with bupropion
Method: single particle / : Zhao Y, Hu T, Yu Z

PDB-8wty: 
Cryo-EM structure of noradrenaline transporter in complex with ziprasidone
Method: single particle / : Zhao Y, Hu T, Yu Z
EMDB-45078: 
E.coli GroEL + PBZ1587 inhibitor
Method: single particle / : Watson ER, Lander GC
EMDB-45079: 
E.coli GroEL apoenzyme
Method: single particle / : Watson ER, Lander GC
EMDB-45080: 
E.Faecium GroEL
Method: single particle / : Watson ER, Lander GC

EMDB-45098: 
E.coli GroEL + PBZ1587 inhibitor C1 reconstruction
Method: single particle / : Watson ER, Lander GC

EMDB-43729: 
Structure of Selenomonas sp. Cascade (SsCascade)
Method: single particle / : Hirano S, Zhang F
EMDB-38758: 
Structural of methylmalonate semialdehyde dehydrogenase ALDH6A1
Method: single particle / : Su G, Xu Y, Luan X
EMDB-36965: 
CryoEM structure of LonC protease hepatmer, apo state
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
EMDB-36966: 
CryoEM structure of LonC protease open hexamer, apo state
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
EMDB-36967: 
CryoEM of LonC open pentamer, apo state
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
EMDB-36968: 
CryoEM structure of LonC heptamer in presence of AGS
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
EMDB-36969: 
CryoEM structure of LonC protease hexamer in presence of AGS
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
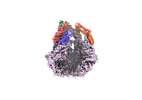
EMDB-36970: 
CryoEM structure of LonC protease open pentamer in presence of AGS
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
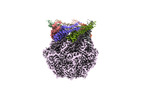
EMDB-36971: 
CryoEM structure of LonC S582A hepatmer with Lysozyme
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
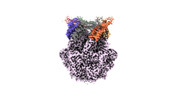
EMDB-36972: 
CryoEM structure of LonC S582A hexamer with Lysozyme
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
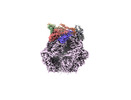
EMDB-36973: 
CryoEM structure of LonC protease S582A open hexamer with lysozyme
Method: single particle / : Li M, Hsieh K, Liu H, Zhang S, Gao Y, Gong Q, Zhang K, Chang C, Li S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search








 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
