+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
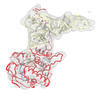
| Title | human ATE1 in complex with Arg-tRNA and a peptide substrate | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | arginylation / ATE1 / complex / TRANSFERASE-RNA complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein arginylation / arginyltransferase / arginyl-tRNA--protein transferase activity / proteasomal protein catabolic process / response to oxidative stress / ubiquitin-dependent protein catabolic process / nucleus / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.7 Å | |||||||||||||||
 Authors Authors | Huang W / Zhang Y / Taylor DJ | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Oligomerization and a distinct tRNA-binding loop are important regulators of human arginyl-transferase function. Authors: Xin Lan / Wei Huang / Su Bin Kim / Dechen Fu / Thilini Abeywansha / Jiemin Lou / Udayakumaran Balamurugan / Yong Tae Kwon / Chang Hoon Ji / Derek J Taylor / Yi Zhang /   Abstract: The arginyl-transferase ATE1 is a tRNA-dependent enzyme that covalently attaches an arginine molecule to a protein substrate. Conserved from yeast to humans, ATE1 deficiency in mice correlates with ...The arginyl-transferase ATE1 is a tRNA-dependent enzyme that covalently attaches an arginine molecule to a protein substrate. Conserved from yeast to humans, ATE1 deficiency in mice correlates with defects in cardiovascular development and angiogenesis and results in embryonic lethality, while conditional knockouts exhibit reproductive, developmental, and neurological deficiencies. Despite the recent revelation of the tRNA binding mechanism and the catalytic cycle of yeast ATE1, the structure-function relationship of ATE1 in higher organisms is not well understood. In this study, we present the three-dimensional structure of human ATE1 in an apo-state and in complex with its tRNA cofactor and a peptide substrate. In contrast to its yeast counterpart, human ATE1 forms a symmetric homodimer, which dissociates upon binding of a substrate. Furthermore, human ATE1 includes a unique and extended loop that wraps around tRNA, creating extensive contacts with the T-arm of the tRNA cofactor. Substituting key residues identified in the substrate binding site of ATE1 abolishes enzymatic activity and results in the accumulation of ATE1 substrates in cells. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42071.map.gz emd_42071.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42071-v30.xml emd-42071-v30.xml emd-42071.xml emd-42071.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42071.png emd_42071.png | 97.6 KB | ||
| Filedesc metadata |  emd-42071.cif.gz emd-42071.cif.gz | 6.6 KB | ||
| Others |  emd_42071_additional_1.map.gz emd_42071_additional_1.map.gz emd_42071_half_map_1.map.gz emd_42071_half_map_1.map.gz emd_42071_half_map_2.map.gz emd_42071_half_map_2.map.gz | 23.1 MB 3.5 MB 3.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42071 http://ftp.pdbj.org/pub/emdb/structures/EMD-42071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42071 | HTTPS FTP |
-Related structure data
| Related structure data |  8uauMC  8tzvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42071.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42071.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Arginyl-tRNA--protein transferase 1/RNA complex
| Entire | Name: Arginyl-tRNA--protein transferase 1/RNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Arginyl-tRNA--protein transferase 1/RNA complex
| Supramolecule | Name: Arginyl-tRNA--protein transferase 1/RNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform ATE1-2 of Arginyl-tRNA--protein transferase 1
| Macromolecule | Name: Isoform ATE1-2 of Arginyl-tRNA--protein transferase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.132898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPAFWAGGSP SVVDYFPSED FYRCGYCKNE SGSRSNGMWA HSMTVQDYQD LIDRGWRRSG KYVYKPVMNQ TCCPQYTIRC RPLQFQPSK SHKKVLKKML KFLAKGEVPK GSCEDEPMDS TMDDAVAGDF ALINKLDIQC DLKTLSDDIK ESLESEGKNS K KEEPQELL ...String: GPAFWAGGSP SVVDYFPSED FYRCGYCKNE SGSRSNGMWA HSMTVQDYQD LIDRGWRRSG KYVYKPVMNQ TCCPQYTIRC RPLQFQPSK SHKKVLKKML KFLAKGEVPK GSCEDEPMDS TMDDAVAGDF ALINKLDIQC DLKTLSDDIK ESLESEGKNS K KEEPQELL QSQDFVGEKL GSGEPSHSVK VHTVPKPGKG ADLSKPPCRK AKEIRKERKR LKLMQQNPAG ELEGFQAQGH PP SLFPPKA KSNQPKSLED LIFESLPENA SHKLEVRLVP VSFEDPEFKS SFSQSFSLYV KYQVAIHQDP PDECGKTEFT RFL CSSPLE AETPPNGPDC GYGSFHQQYW LDGKIIAVGV IDILPNCVSS VYLYYDPDYS FLSLGVYSAL REIAFTRQLH EKTS QLSYY YMGFYIHSCP KMKYKGQYRP SDLLCPETYV WVPIEQCLPS LENSKYCRFN QDPEAVDEDR STEPDRLQVF HKRAI MPYG VYKKQQKDPS EEAAVLQYAS LVGQKCSERM LLFRN UniProtKB: Arginyl-tRNA--protein transferase 1 |
-Macromolecule #2: substrate
| Macromolecule | Name: substrate / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.188244 KDa |
| Sequence | String: DDIAALVVDN GS |
-Macromolecule #3: RNA (76-MER)
| Macromolecule | Name: RNA (76-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.456488 KDa |
| Sequence | String: GGCUCUGUGG CGCAAUGGAU AGCGCAUUGG ACUUCUAAUU CAAAGGUUGU GGGUUCGAAU CCCACCAGAG UCGCCA |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















