[English] 日本語
 Yorodumi
Yorodumi- EMDB-38758: Structural of methylmalonate semialdehyde dehydrogenase ALDH6A1 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural of methylmalonate semialdehyde dehydrogenase ALDH6A1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dehydrogenase / ALDH6A1 / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationvaline metabolic process / thymine metabolic process / malonate-semialdehyde dehydrogenase (acetylating) activity / methylmalonate-semialdehyde dehydrogenase (CoA-acylating) / methylmalonate-semialdehyde dehydrogenase (acylating, NAD) activity / thymine catabolic process / L-valine catabolic process / branched-chain amino acid catabolic process / Branched-chain amino acid catabolism / brown fat cell differentiation ...valine metabolic process / thymine metabolic process / malonate-semialdehyde dehydrogenase (acetylating) activity / methylmalonate-semialdehyde dehydrogenase (CoA-acylating) / methylmalonate-semialdehyde dehydrogenase (acylating, NAD) activity / thymine catabolic process / L-valine catabolic process / branched-chain amino acid catabolic process / Branched-chain amino acid catabolism / brown fat cell differentiation / mitochondrial matrix / mitochondrion / RNA binding / nucleoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.75 Å | |||||||||
 Authors Authors | Su G / Xu Y / Luan X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Medicine Plus / Year: 2024 Journal: Medicine Plus / Year: 2024Title: Structural and biochemical basis of methylmalonate semialdehyde dehydrogenase ALDH6A1 Authors: Su G / Ju K / Xu Y / Jin Y / Chen L / Zhang S / Luan X | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38758.map.gz emd_38758.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38758-v30.xml emd-38758-v30.xml emd-38758.xml emd-38758.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38758.png emd_38758.png | 48.9 KB | ||
| Filedesc metadata |  emd-38758.cif.gz emd-38758.cif.gz | 5.8 KB | ||
| Others |  emd_38758_half_map_1.map.gz emd_38758_half_map_1.map.gz emd_38758_half_map_2.map.gz emd_38758_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38758 http://ftp.pdbj.org/pub/emdb/structures/EMD-38758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38758 | HTTPS FTP |
-Related structure data
| Related structure data |  8xxqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38758.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38758.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
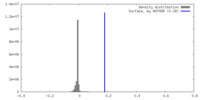
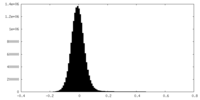
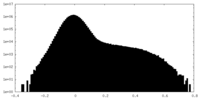
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38758_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
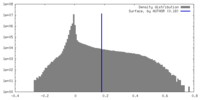
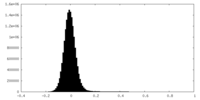
| Density Histograms |
-Half map: #2
| File | emd_38758_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
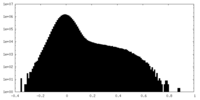
| Density Histograms |
- Sample components
Sample components
-Entire : ALDH6A1
| Entire | Name: ALDH6A1 |
|---|---|
| Components |
|
-Supramolecule #1: ALDH6A1
| Supramolecule | Name: ALDH6A1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Methylmalonate-semialdehyde/malonate-semialdehyde dehydrogenase [...
| Macromolecule | Name: Methylmalonate-semialdehyde/malonate-semialdehyde dehydrogenase [acylating], mitochondrial type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO EC number: methylmalonate-semialdehyde dehydrogenase (CoA-acylating) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.146129 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SSVPTVKLFI GGKFVESKSD KWIDIHNPAT NEVIGRVPQA TKAEMDAAIA SCKRAFPAWA DTSVLSRQQV LLRYQQLIKE NLKEIAKLI TLEQGKTLAD AEGDVFRGLQ VVEHACSVTS LMMGETMPSI TKDMDLYSYR LPLGVCAGIA PFNFPAMIPL W MFPMAMVC ...String: SSVPTVKLFI GGKFVESKSD KWIDIHNPAT NEVIGRVPQA TKAEMDAAIA SCKRAFPAWA DTSVLSRQQV LLRYQQLIKE NLKEIAKLI TLEQGKTLAD AEGDVFRGLQ VVEHACSVTS LMMGETMPSI TKDMDLYSYR LPLGVCAGIA PFNFPAMIPL W MFPMAMVC GNTFLMKPSE RVPGATMLLA KLLQDSGAPD GTLNIIHGQH EAVNFICDHP DIKAISFVGS NKAGEYIFER GS RHGKRVQ ANMGAKNHGV VMPDANKENT LNQLVGAAFG AAGQRCMALS TAVLVGEAKK WLPELVEHAK NLRVNAGDQP GAD LGPLIT PQAKERVCNL IDSGTKEGAS ILLDGRKIKV KGYENGNFVG PTIISNVKPN MTCYKEEIFG PVLVVLETET LDEA IQIVN NNPYGNGTAI FTTNGATARK YAHLVDVGQV GVNVPIPVPL PMFSFTGSRS SFRGDTNFYG KQGIQFYTQL KTITS QWKE EDATLSSPAV VMPTM UniProtKB: Methylmalonate-semialdehyde/malonate-semialdehyde dehydrogenase [acylating], mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)