-Search query
-Search result
Showing 1 - 50 of 616 items for (author: kay & le)

EMDB-44123: 
Cryo-EM density of GluK2 amino-terminal domain (GluK2-ATD) from the open-state structure of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to ConA
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-44126: 
Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-44127: 
Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-17298: 
Cryo-EM structure of Phthaloyl-CoA decarboxylase (Pcd) bound with substrate analog/inhibitor, 2-CN-benzoyl-CoA
Method: single particle / : Kayastha K, Ermler U

PDB-8oz5: 
Cryo-EM structure of Phthaloyl-CoA decarboxylase (Pcd) bound with substrate analog/inhibitor, 2-CN-benzoyl-CoA
Method: single particle / : Kayastha K, Ermler U

EMDB-17324: 
Cryo-EM structure of Phthaloyl-CoA decarboxylase (Pcd) bound with product, benzoyl-CoA
Method: single particle / : Kayastha K, Ermler U

PDB-8p02: 
Cryo-EM structure of Phthaloyl-CoA decarboxylase (Pcd) bound with product, benzoyl-CoA
Method: single particle / : Kayastha K, Ermler U

EMDB-19066: 
TREK2 in OGNG/CHS detergent micelle with biparatopic inhibitory nanobody Nb6158
Method: single particle / : Smith KHM, Tucker SJ

EMDB-44124: 
Structure of concanavalin A (ConA) dimer from the open-state structure of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one ConA dimer. Type II interface between GluK2 ligand-binding domain and ConA
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-44125: 
Structure of concanavalin A (ConA) dimer from the open-state structure of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two ConA dimers. Type I interface between GluK2 ligand-binding domain and ConA
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-44128: 
Ligand-binding and transmembrane domains of kainate receptor GluK2 in the open state, a complex with agonist glutamate and positive allosteric modulator BPAM344
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-44129: 
Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers. Composite map.
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-44130: 
Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer. Composite map.
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-44131: 
Kainate receptor GluK2 in complex with agonist glutamate with pseudo 4-fold symmetrical ligand-binding domain layer
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-44132: 
Kainate receptor GluK2 in complex with agonist glutamate with asymmetric ligand-binding domain layer
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

PDB-9b33: 
Structure of concanavalin A (ConA) dimer from the open-state structure of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one ConA dimer. Type II interface between GluK2 ligand-binding domain and ConA
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

PDB-9b34: 
Structure of concanavalin A (ConA) dimer from the open-state structure of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two ConA dimers. Type I interface between GluK2 ligand-binding domain and ConA
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

PDB-9b35: 
Ligand-binding and transmembrane domains of kainate receptor GluK2 in the open state, a complex with agonist glutamate and positive allosteric modulator BPAM344
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

PDB-9b36: 
Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers. Composite map.
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

PDB-9b37: 
Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer. Composite map.
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

PDB-9b38: 
Kainate receptor GluK2 in complex with agonist glutamate with pseudo 4-fold symmetrical ligand-binding domain layer
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

PDB-9b39: 
Kainate receptor GluK2 in complex with agonist glutamate with asymmetric ligand-binding domain layer
Method: single particle / : Nadezhdin KD, Gangwar SP, Sobolevsky AI

EMDB-37963: 
Cryo-EM structure of the hamster prion 23-144 fibril at pH 3.7
Method: helical / : Lee CH, Saw JE, Chen E, Wang CH, Chen R

PDB-8wzx: 
Cryo-EM structure of the hamster prion 23-144 fibril at pH 3.7
Method: helical / : Lee CH, Saw JE, Chen E, Wang CH, Chen R
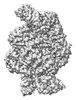
EMDB-18963: 
Structure of the SFTSV L protein in a transcription-priming state without capped RNA [TRANSCRIPTION-PRIMING (in vitro)]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M
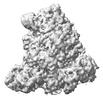
EMDB-18967: 
Structure of the SFTSV L protein in a transcription-priming state with bound capped RNA [TRANSCRIPTION-PRIMING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-18969: 
Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8r6u: 
Structure of the SFTSV L protein in a transcription-priming state without capped RNA [TRANSCRIPTION-PRIMING (in vitro)]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8r6w: 
Structure of the SFTSV L protein in a transcription-priming state with bound capped RNA [TRANSCRIPTION-PRIMING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8r6y: 
Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-42994: 
Apo-state cryo-EM structure of human TRPV3 in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI
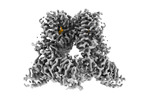
EMDB-42995: 
Open-state cryo-EM structure of human TRPV3 in presence of tetrahydrocannabivarin (THCV) in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

EMDB-42996: 
Inactivated-state cryo-EM structure of human TRPV3 in presence of tetrahydrocannabivarin (THCV) in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

EMDB-42997: 
Open-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

EMDB-42998: 
Inactivated-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

PDB-8v6k: 
Apo-state cryo-EM structure of human TRPV3 in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

PDB-8v6l: 
Open-state cryo-EM structure of human TRPV3 in presence of tetrahydrocannabivarin (THCV) in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

PDB-8v6m: 
Inactivated-state cryo-EM structure of human TRPV3 in presence of tetrahydrocannabivarin (THCV) in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

PDB-8v6n: 
Open-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

PDB-8v6o: 
Inactivated-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

EMDB-19276: 
Cryo-EM of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1)
Method: single particle / : Wiseman B, Nishio S, Jovine L

EMDB-19277: 
Cryo-EM of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C2)
Method: single particle / : Wiseman B, Nishio S, Jovine L
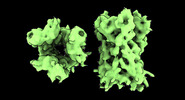
EMDB-17704: 
Subtomogram average of Vaccinia A10 trimer with open center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J
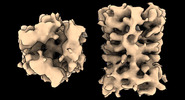
EMDB-17708: 
Subtomogram average of Vaccinia A10 trimer with tight center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-17753: 
Subtomogram average of Vaccinia A10 trimer from in situ cores
Method: subtomogram averaging / : Turonova B, Liu J
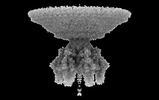
EMDB-41649: 
P22 Mature Virion tail - C6 Localized Reconstruction
Method: single particle / : Iglesias S, Cingolani G, Feng-Hou C

EMDB-41651: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution
Method: single particle / : Iglesias SM, Cingolani G, Feng-Hou C

EMDB-41819: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution
Method: single particle / : Iglesias SM, Feng-Hou C, Cingolani G

PDB-8tvr: 
In situ cryo-EM structure of bacteriophage P22 tail hub protein: tailspike protein complex at 2.8A resolution
Method: single particle / : Iglesias S, Cingolani G, Feng-Hou C

PDB-8tvu: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution
Method: single particle / : Iglesias SM, Cingolani G, Feng-Hou C
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
