-Search query
-Search result
Showing 1 - 50 of 2,537 items for (author: fan & q)

EMDB-37754: 
Fe-O nanocluster of form-IX in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-37755: 
Fe-O nanocluster of form-VIII in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-37757: 
Fe-O nanocluster of form-X in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-37758: 
Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-37759: 
Fe-O nanocluster of form-XII in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wqu: 
Fe-O nanocluster of form-IX in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wqv: 
Fe-O nanocluster of form-VIII in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wqx: 
Fe-O nanocluster of form-X in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wqy: 
Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wr0: 
Fe-O nanocluster of form-XII in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-36599: 
Structure of E6AP-E6 complex in Att1 state
Method: single particle / : Wang Z, Yu X

EMDB-36600: 
Structure of E6AP-E6 complex in Att2 state
Method: single particle / : Wang Z, Yu X

EMDB-36601: 
Structure of E6AP-E6 complex in Att3 state
Method: single particle / : Wang Z, Yu X

EMDB-36602: 
Structure of E6AP-E6 complex in Det1 state
Method: single particle / : Wang Z, Yu X

EMDB-36603: 
Structure of E6AP-E6 complex in Det2 state
Method: single particle / : Wang Z, Yu X

EMDB-36604: 
Structure of human full-length E6AP
Method: single particle / : Wang Z, Yu X

EMDB-36672: 
Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab
Method: single particle / : Liu B, Liu HH, Han P, Qi JX

PDB-8jva: 
Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab
Method: single particle / : Liu B, Liu HH, Han P, Qi JX

EMDB-36907: 
Cryo-EM structure of the RC-LH core comples from Halorhodospira halochloris
Method: single particle / : Wang GL, Qi CH, Yu LJ

PDB-8k5o: 
Cryo-EM structure of the RC-LH core comples from Halorhodospira halochloris
Method: single particle / : Wang GL, Qi CH, Yu LJ

EMDB-18818: 
Structure of avian H5N1 influenza A polymerase dimer in complex with human ANP32B.
Method: single particle / : Carrique L, Staller E, Keown JR, Fan H, Fodor E, Grimes JM

EMDB-18819: 
Structure of avian H5N1 influenza A polymerase dimer in complex with human ANP32B (Consensus map)
Method: single particle / : Carrique L, Staller E, Keown JR, Fan H, Fodor E, Grimes JM

EMDB-18820: 
Structure of avian H5N1 influenza A polymerase dimer in complex with human ANP32B (Encapsidase focused).
Method: single particle / : Carrique L, Staller E, Keown JR, Fan H, Fodor E, Grimes JM

EMDB-18821: 
Structure of avian H5N1 influenza A polymerase dimer in complex with human ANP32B (Replicase focused).
Method: single particle / : Carrique L, Staller E, Keown JR, Fan H, Fodor E, Grimes JM

EMDB-18822: 
Structure of avian H5N1 influenza A polymerase in complex with human ANP32B.
Method: single particle / : Carrique L, Staller E, Keown JR, Fan H, Fodor E, Grimes JM

PDB-8r1j: 
Structure of avian H5N1 influenza A polymerase dimer in complex with human ANP32B.
Method: single particle / : Carrique L, Staller E, Keown JR, Fan H, Fodor E, Grimes JM

PDB-8r1l: 
Structure of avian H5N1 influenza A polymerase in complex with human ANP32B.
Method: single particle / : Carrique L, Staller E, Keown JR, Fan H, Fodor E, Grimes JM

EMDB-36732: 
Cryo-EM structure of the gasdermin pore from Trichoplax adhaerens
Method: single particle / : Hou YJ, Sun Q, Zeng H, Ding J

EMDB-36733: 
Cryo-EM structure of the gasdermin pore from Trichoplax adhaerens
Method: single particle / : Hou YJ, Sun Q, Zeng H, Ding J

EMDB-36734: 
Cryo-EM structure of RCD-1 pore from Neurospora crassa
Method: single particle / : Hou YJ, Sun Q, Li Y, Ding J

PDB-8jyw: 
Cryo-EM structure of the gasdermin pore from Trichoplax adhaerens
Method: single particle / : Hou YJ, Sun Q, Zeng H, Ding J

PDB-8jyz: 
Cryo-EM structure of RCD-1 pore from Neurospora crassa
Method: single particle / : Hou YJ, Sun Q, Li Y, Ding J

EMDB-36849: 
Nipah virus Attachment glycoprotein with 41-6 antibody fragment
Method: single particle / : Sun MM

PDB-8k3c: 
Nipah virus Attachment glycoprotein with 41-6 antibody fragment
Method: single particle / : Sun MM
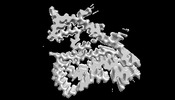
EMDB-36045: 
The cryo-EM structure of PiB bound TMEM106B fibril.
Method: helical / : Zhao QY, Tao YQ, Yan F, Liu C, Li D
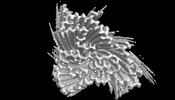
EMDB-36043: 
The cryo-EM structure of Fe3+ induced alpha-syn fibril.
Method: helical / : Zhao QY, Tao YQ, Yan F, Liu C, Li D

EMDB-43991: 
Cryo-EM structure of apo state human Cav3.2
Method: single particle / : Fan X, Huang J, Yan N

EMDB-43992: 
Cryo-EM structure of human Cav3.2 with TTA-A2
Method: single particle / : Fan X, Huang J, Yan N

EMDB-43993: 
Cryo-EM structure of human Cav3.2 with TTA-P2
Method: single particle / : Fan X, Huang J, Yan N

EMDB-43994: 
Cryo-EM structure of human Cav3.2 with ML218
Method: single particle / : Fan X, Huang J, Yan N

EMDB-43995: 
Cryo-EM structure of human Cav3.2 with ACT-709478
Method: single particle / : Fan X, Huang J, Yan N

PDB-9ayg: 
Cryo-EM structure of apo state human Cav3.2
Method: single particle / : Fan X, Huang J, Yan N

PDB-9ayh: 
Cryo-EM structure of human Cav3.2 with TTA-A2
Method: single particle / : Fan X, Huang J, Yan N

PDB-9ayj: 
Cryo-EM structure of human Cav3.2 with TTA-P2
Method: single particle / : Fan X, Huang J, Yan N

PDB-9ayk: 
Cryo-EM structure of human Cav3.2 with ML218
Method: single particle / : Fan X, Huang J, Yan N
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search








 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
