+Search query
-Structure paper
| Title | Structural insights into the functional mechanism of the ubiquitin ligase E6AP. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 3531, Year 2024 |
| Publish date | Apr 26, 2024 |
 Authors Authors | Zhen Wang / Fengying Fan / Zhihai Li / Fei Ye / Qingxia Wang / Rongchao Gao / Jiaxuan Qiu / Yixin Lv / Min Lin / Wenwen Xu / Cheng Luo / Xuekui Yu /  |
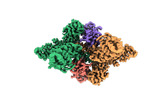
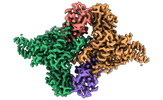
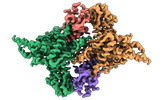
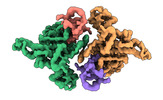
| PubMed Abstract | E6AP dysfunction is associated with Angelman syndrome and Autism spectrum disorder. Additionally, the host E6AP is hijacked by the high-risk HPV E6 to aberrantly ubiquitinate the tumor suppressor ...E6AP dysfunction is associated with Angelman syndrome and Autism spectrum disorder. Additionally, the host E6AP is hijacked by the high-risk HPV E6 to aberrantly ubiquitinate the tumor suppressor p53, which is linked with development of multiple types of cancer, including most cervical cancers. Here we show that E6AP and the E6AP/E6 complex exist, respectively, as a monomer and a dimer of the E6AP/E6 protomer. The short α1-helix of E6AP transforms into a longer helical structure when in complex with E6. The extended α1-helices of the dimer intersect symmetrically and contribute to the dimerization. The two protomers sway around the crossed region of the two α1-helices to promote the attachment and detachment of substrates to the catalytic C-lobe of E6AP, thus facilitating ubiquitin transfer. These findings, complemented by mutagenesis analysis, suggest that the α1-helix, through conformational transformations, controls the transition between the inactive monomer and the active dimer of E6AP. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:38670961 / PubMed:38670961 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.6 - 5.1 Å |
| Structure data | EMDB-36599, PDB-8jrn: EMDB-36600, PDB-8jro: EMDB-36601, PDB-8jrp: EMDB-36602, PDB-8jrq: EMDB-36603, PDB-8jrr:  EMDB-36604: Structure of human full-length E6AP |
| Chemicals |  ChemComp-ZN: |
| Source |
|
 Keywords Keywords | LIGASE/ONCOPROTEIN / Complex / Viral protein / Tumor / LIGASE-ONCOPROTEIN complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








 homo sapiens (human)
homo sapiens (human) human papillomavirus 16
human papillomavirus 16