-Search query
-Search result
Showing all 18 items for (author: bhakta & s)

EMDB-30598: 
Cryo-EM structure of 70S ribosome in complex with peptide deformylase and trigger factor
Method: single particle / : Akbar S, Bhakta S, Sengupta J

EMDB-30611: 
Cryo-EM map of 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase
Method: single particle / : Akbar S, Bhakta S

PDB-7d6z: 
Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase and trigger factor
Method: single particle / : Akbar S, Bhakta S, Sengupta J

PDB-7d80: 
Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase
Method: single particle / : Akbar S, Bhakta S, Sengupta J
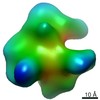
EMDB-9878: 
Cryo EM density map of Resveratrol-stabilized bioactive insulin oligomer
Method: single particle / : Sengupta J, Pathak BK

PDB-6jr3: 
Crystal structure of insulin hexamer fitted into cryo EM density map where each dimer was kept as rigid body
Method: single particle / : Sengupta J, Pathak BK, Bhakta S

EMDB-9750: 
Cryo-EM density map of E. coli 70S ribosome in complex with peptide deformylase enzyme
Method: single particle / : Sengupta J, Akbar S

EMDB-9752: 
Cryo-EM density map of E. coli 70S ribosome in complex with methionine aminopeptidase enzyme
Method: single particle / : Sengupta J, Bhakta S

EMDB-9753: 
Cryo-EM density map of peptide deformylase and methionine aminopeptidase bound to the E. coli 70S ribosome
Method: single particle / : Sengupta J, Bhakta S

EMDB-9759: 
Cryo-EM density map of methionine aminopeptidase enzyme and chaperone trigger factor bound to the E. coli 70S ribosome
Method: single particle / : Sengupta J, Bhakta S

EMDB-9778: 
Cryo-EM density map of peptide deformylase enzyme and chaperone trigger factor bound to the E. coli 70S ribosome
Method: single particle / : Sengupta J, Bhakta S

PDB-6iy7: 
E. coli peptide deformylase crystal structure fitted into the cryo-EM density map of E. coli 70S ribosome in complex with peptide deformylase
Method: single particle / : Sengupta J, Akbar S, Bhakta S

PDB-6iz7: 
E. coli methionine aminopeptidase crystal structure fitted into the cryo-EM density map of E. coli 70S ribosome in complex with methionine aminopeptidase
Method: single particle / : Sengupta J, Bhakta S, Akbar S

PDB-6izi: 
Crystal structure of E. coli peptide deformylase and methionine aminopeptidase fitted into the cryo-EM density map of the complex
Method: single particle / : Sengupta J, Bhakta S, Akbar S

PDB-6j0a: 
Crystal structure of E. coli methionine aminopeptidase enzyme and chaperone trigger factor fitted into the cryo-EM density map of the complex
Method: single particle / : Sengupta J, Bhakta S, Akbar S

PDB-6j45: 
Crystal structure of E. coli peptide deformylase enzyme and chaperone trigger factor fitted into the cryo-EM density map of the complex
Method: single particle / : Sengupta J, Bhakta S, Akbar S
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
