+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10618 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
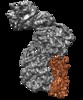
| Title | The human core BBSome complex (BBS 1,4,5,8,9,18) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ciliary transport / Arl6 effector / adaptor protein / complex / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase II-specific DNA-binding transcription factor binding => GO:0061629 / BBSome / melanosome transport / motile cilium assembly / BBSome-mediated cargo-targeting to cilium / response to stimulus / phosphatidylinositol-3-phosphate binding / ciliary membrane / heart looping / axoneme ...RNA polymerase II-specific DNA-binding transcription factor binding => GO:0061629 / BBSome / melanosome transport / motile cilium assembly / BBSome-mediated cargo-targeting to cilium / response to stimulus / phosphatidylinositol-3-phosphate binding / ciliary membrane / heart looping / axoneme / centriolar satellite / cilium assembly / intracellular transport / visual perception / ciliary basal body / protein transport / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Klink BU / Raunser S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structure of the human BBSome core complex. Authors: Björn Udo Klink / Christos Gatsogiannis / Oliver Hofnagel / Alfred Wittinghofer / Stefan Raunser /  Abstract: The BBSome is a heterooctameric protein complex that plays a central role in primary cilia homeostasis. Its malfunction causes the severe ciliopathy Bardet-Biedl syndrome (BBS). The complex acts as a ...The BBSome is a heterooctameric protein complex that plays a central role in primary cilia homeostasis. Its malfunction causes the severe ciliopathy Bardet-Biedl syndrome (BBS). The complex acts as a cargo adapter that recognizes signaling proteins such as GPCRs and links them to the intraflagellar transport machinery. The underlying mechanism is poorly understood. Here we present a high-resolution cryo-EM structure of a human heterohexameric core subcomplex of the BBSome. The structure reveals the architecture of the complex in atomic detail. It explains how the subunits interact with each other and how disease-causing mutations hamper this interaction. The complex adopts a conformation that is open for binding to membrane-associated GTPase Arl6 and a large positively charged patch likely strengthens the interaction with the membrane. A prominent negatively charged cleft at the center of the complex is likely involved in binding of positively charged signaling sequences of cargo proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10618.map.gz emd_10618.map.gz | 4.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10618-v30.xml emd-10618-v30.xml emd-10618.xml emd-10618.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10618.png emd_10618.png | 129.9 KB | ||
| Filedesc metadata |  emd-10618.cif.gz emd-10618.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10618 http://ftp.pdbj.org/pub/emdb/structures/EMD-10618 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10618 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10618 | HTTPS FTP |
-Validation report
| Summary document |  emd_10618_validation.pdf.gz emd_10618_validation.pdf.gz | 212.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10618_full_validation.pdf.gz emd_10618_full_validation.pdf.gz | 211.9 KB | Display | |
| Data in XML |  emd_10618_validation.xml.gz emd_10618_validation.xml.gz | 6.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10618 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10618 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10618 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10618 | HTTPS FTP |
-Related structure data
| Related structure data |  6xtbMC  6xt9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10618.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10618.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human BBSome core complex
| Entire | Name: Human BBSome core complex |
|---|---|
| Components |
|
-Supramolecule #1: Human BBSome core complex
| Supramolecule | Name: Human BBSome core complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: BBSome core complex containing BBS1,4,5,8,9 and 18. Only BBS5 was modelled into this map since we obtained another map with higher resolution for the other subunits (see related entry) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Bardet-Biedl syndrome 5 protein
| Macromolecule | Name: Bardet-Biedl syndrome 5 protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.797926 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSVLDALWED RDVRFDLSAQ QMKTRPGEVL IDCLDSIEDT KGNNGDRGRL LVTNLRILWH SLALSRVNVS VGYNCILNIT TRTANSKLR GQTEALYILT KCNSTRFEFI FTNLVPGSPR LFTSVMAVHR AYETSKMYRD FKLRSALIQN KQLRLLPQEH V YDKINGVW ...String: MSVLDALWED RDVRFDLSAQ QMKTRPGEVL IDCLDSIEDT KGNNGDRGRL LVTNLRILWH SLALSRVNVS VGYNCILNIT TRTANSKLR GQTEALYILT KCNSTRFEFI FTNLVPGSPR LFTSVMAVHR AYETSKMYRD FKLRSALIQN KQLRLLPQEH V YDKINGVW NLSSDQGNLG TFFITNVRIV WHANMNDSFN VSIPYLQIRS IKIRDSKFGL ALVIESSQQS GGYVLGFKID PV EKLQESV KEINSLHKVY SASPIFGVDY EMEEKPQPLE ALTVEQIQDD VEIDSDGHTD AFVAYFADGN KQQDREPVFS EEL GLAIEK LKDGFTLQGL WEVMS UniProtKB: Bardet-Biedl syndrome 5 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 286 K / Instrument: FEI VITROBOT MARK III Details: double blot with 2 minutes incubation after first sample application. | ||||||||
| Details | The sample was cross linked with 0.5% glutaraldehyde |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-50 / Number real images: 15266 / Average exposure time: 15.0 sec. / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 1.0 µm / Calibrated defocus min: 0.3 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller