+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10617 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
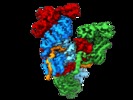
| Title | Subunits BBS 1,4,8,9,18 of the human BBSome complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ciliary transport / Arl6 effector / adaptor protein / complex / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of non-motile cilium assembly / protein localization to photoreceptor outer segment / receptor localization to non-motile cilium / BBSome / photoreceptor cell morphogenesis / retinal rod cell development / sperm flagellum assembly / negative regulation of appetite by leptin-mediated signaling pathway / photoreceptor cell outer segment organization / smoothened binding ...regulation of non-motile cilium assembly / protein localization to photoreceptor outer segment / receptor localization to non-motile cilium / BBSome / photoreceptor cell morphogenesis / retinal rod cell development / sperm flagellum assembly / negative regulation of appetite by leptin-mediated signaling pathway / photoreceptor cell outer segment organization / smoothened binding / olfactory behavior / regulation of cilium beat frequency involved in ciliary motility / microtubule anchoring at centrosome / sensory processing / photoreceptor connecting cilium / negative regulation of actin filament polymerization / patched binding / BBSome-mediated cargo-targeting to cilium / ventricular system development / positive regulation of cilium assembly / striatum development / protein localization to cilium / melanosome transport / sensory perception / Golgi to plasma membrane protein transport / maintenance of protein location in nucleus / positive regulation of multicellular organism growth / regulation of stress fiber assembly / non-motile cilium assembly / brain morphogenesis / photoreceptor cell maintenance / retina homeostasis / perinuclear theca / negative regulation of systemic arterial blood pressure / hormone metabolic process / cartilage development / fertilization / protein localization to centrosome / non-motile cilium / ciliary transition zone / centrosome cycle / neural precursor cell proliferation / fat pad development / eating behavior / motile cilium / ciliary membrane / erythrocyte homeostasis / heart looping / face development / pericentriolar material / dendrite development / B cell homeostasis / adult behavior / mitotic cytokinesis / dynactin binding / regulation of lipid metabolic process / cilium assembly / spermatid development / social behavior / photoreceptor outer segment / axoneme / anatomical structure morphogenesis / alpha-tubulin binding / fat cell differentiation / beta-tubulin binding / photoreceptor inner segment / visual perception / acrosomal vesicle / response to endoplasmic reticulum stress / regulation of cytokinesis / centriole / hippocampus development / lipid metabolic process / neural tube closure / phosphoprotein binding / cerebral cortex development / microtubule cytoskeleton organization / centriolar satellite / neuron migration / Wnt signaling pathway / fibrillar center / sensory perception of smell / sperm principal piece / protein transport / sperm midpiece / retina development in camera-type eye / gene expression / protein-macromolecule adaptor activity / RNA polymerase II-specific DNA-binding transcription factor binding / cilium / ciliary basal body / negative regulation of gene expression / centrosome / nucleoplasm / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Klink BU / Raunser S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structure of the human BBSome core complex. Authors: Björn Udo Klink / Christos Gatsogiannis / Oliver Hofnagel / Alfred Wittinghofer / Stefan Raunser /  Abstract: The BBSome is a heterooctameric protein complex that plays a central role in primary cilia homeostasis. Its malfunction causes the severe ciliopathy Bardet-Biedl syndrome (BBS). The complex acts as a ...The BBSome is a heterooctameric protein complex that plays a central role in primary cilia homeostasis. Its malfunction causes the severe ciliopathy Bardet-Biedl syndrome (BBS). The complex acts as a cargo adapter that recognizes signaling proteins such as GPCRs and links them to the intraflagellar transport machinery. The underlying mechanism is poorly understood. Here we present a high-resolution cryo-EM structure of a human heterohexameric core subcomplex of the BBSome. The structure reveals the architecture of the complex in atomic detail. It explains how the subunits interact with each other and how disease-causing mutations hamper this interaction. The complex adopts a conformation that is open for binding to membrane-associated GTPase Arl6 and a large positively charged patch likely strengthens the interaction with the membrane. A prominent negatively charged cleft at the center of the complex is likely involved in binding of positively charged signaling sequences of cargo proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10617.map.gz emd_10617.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10617-v30.xml emd-10617-v30.xml emd-10617.xml emd-10617.xml | 23.9 KB 23.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10617.png emd_10617.png | 166.1 KB | ||
| Masks |  emd_10617_msk_1.map emd_10617_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10617.cif.gz emd-10617.cif.gz | 8 KB | ||
| Others |  emd_10617_half_map_1.map.gz emd_10617_half_map_1.map.gz emd_10617_half_map_2.map.gz emd_10617_half_map_2.map.gz | 39.4 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10617 http://ftp.pdbj.org/pub/emdb/structures/EMD-10617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10617 | HTTPS FTP |
-Related structure data
| Related structure data |  6xt9MC  6xtbC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10617.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10617.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10617_msk_1.map emd_10617_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10617_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10617_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BBSome core complex
| Entire | Name: BBSome core complex |
|---|---|
| Components |
|
-Supramolecule #1: BBSome core complex
| Supramolecule | Name: BBSome core complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: BBSome core complex containing BBS1,4, 8, 9 and 18. BBS5 was also present in the sample preparation, but was only visible in a subset of particles (see related entry) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Bardet-Biedl syndrome 1 protein
| Macromolecule | Name: Bardet-Biedl syndrome 1 protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 65.159266 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAASSSDSD ACGAESNEAN SKWLDAHYDP MANIHTFSAC LALADLHGDG EYKLVVGDLG PGGQQPRLKV LKGPLVMTES PLPALPAAA ATFLMEQHEP RTPALALASG PCVYVYKNLR PYFKFSLPQL PPNPLEQDLW NQAKEDRIDP LTLKEMLESI R ETAEEPLS ...String: MAAASSSDSD ACGAESNEAN SKWLDAHYDP MANIHTFSAC LALADLHGDG EYKLVVGDLG PGGQQPRLKV LKGPLVMTES PLPALPAAA ATFLMEQHEP RTPALALASG PCVYVYKNLR PYFKFSLPQL PPNPLEQDLW NQAKEDRIDP LTLKEMLESI R ETAEEPLS IQSLRFLQLE LSEMEAFVNQ HKSNSIKRQT VITTMTTLKK NLADEDAVSC LVLGTENKEL LVLDPEAFTI LA KMSLPSV PVFLEVSGQF DVEFRLAAAC RNGNIYILRR DSKHPKYCIE LSAQPVGLIR VHKVLVVGST QDSLHGFTHK GKK LWTVQM PAAILTMNLL EQHSRGLQAV MAGLANGEVR IYRDKALLNV IHTPDAVTSL CFGRYGREDN TLIMTTRGGG LIIK ILKRT AVFVEGGSEV GPPPAQAMKL NVPRKTRLYV DQTLREREAG TAMHRAFQTD LYLLRLRAAR AYLQALESSL SPLST TARE PLKLHAVVQG LGPTFKLTLH LQNTSTTRPV LGLLVCFLYN EALYSLPRAF FKVPLLVPGL NYPLETFVES LSNKGI SDI IKVLVLREGQ SAPLLSAHVN MPGSEGLAAA UniProtKB: BBSome complex member BBS1 |
-Macromolecule #2: Bardet-Biedl syndrome 4 protein
| Macromolecule | Name: Bardet-Biedl syndrome 4 protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.46302 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHGPM AEERVATRTQ FPVSTESQKP RQKKAPEFPI LEKQNWLIHL HYIRKDYEAC KAVIKEQLQE TQGLCEYAIY VQALIFRLE GNIQESLELF QTCAVLSPQS ADNLKQVARS LFLLGKHKAA IEVYNEAAKL NQKDWEISHN LGVCYIYLKQ F NKAQDQLH ...String: MHHHHHHGPM AEERVATRTQ FPVSTESQKP RQKKAPEFPI LEKQNWLIHL HYIRKDYEAC KAVIKEQLQE TQGLCEYAIY VQALIFRLE GNIQESLELF QTCAVLSPQS ADNLKQVARS LFLLGKHKAA IEVYNEAAKL NQKDWEISHN LGVCYIYLKQ F NKAQDQLH NALNLNRHDL TYIMLGKIHL LEGDLDKAIE VYKKAVEFSP ENTELLTTLG LLYLQLGIYQ KAFEHLGNAL TY DPTNYKA ILAAGSMMQT HGDFDVALTK YRVVACAVPE SPPLWNNIGM CFFGKKKYVA AISCLKRANY LAPFDWKILY NLG LVHLTM QQYASAFHFL SAAINFQPKM GELYMLLAVA LTNLEDIENA KRAYAEAVHL DKCNPLVNLN YAVLLYNQGE KKNA LAQYQ EMEKKVSLLK DNSSLEFDSE MVEMAQKLGA ALQVGEALVW TKPVKDPKSK HQTTSTSKPA SFQQPLGSNQ ALGQA MSSA AAYRTLPSGA GGTSQFTKPP SLPLEPEPAV ESSPTETSEQ IREK UniProtKB: BBSome complex member BBS4 |
-Macromolecule #3: Tetratricopeptide repeat domain 8 isoform 2
| Macromolecule | Name: Tetratricopeptide repeat domain 8 isoform 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.702539 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDYKDDDDKA GPMSSEMEPL LLAWSYFRRR KFQLCADLCT QMLEKSPYDQ AAWILKARAL TEMVYIDEID VDQEGIAEMM LDENAIAQV PRPGTSLKLP GTNQTGGPSQ AVRPITQAGR PITGFLRPST QSGRPGTMEQ AIRTPRTAYT ARPITSSSGR F VRLGTASM ...String: MDYKDDDDKA GPMSSEMEPL LLAWSYFRRR KFQLCADLCT QMLEKSPYDQ AAWILKARAL TEMVYIDEID VDQEGIAEMM LDENAIAQV PRPGTSLKLP GTNQTGGPSQ AVRPITQAGR PITGFLRPST QSGRPGTMEQ AIRTPRTAYT ARPITSSSGR F VRLGTASM LTSPDGPFIN LSRLNLTKYS QKPKLAKALF EYIFHHENDV KTALDLAALS TEHSQYKDWW WKVQIGKCYY RL GMYREAE KQFKSALKQQ EMVDTFLYLA KVYVSLDQPV TALNLFKQGL DKFPGEVTLL CGIARIYEEM NNMSSAAEYY KEV LKQDNT HVEAIACIGS NHFYSDQPEI ALRFYRRLLQ MGIYNGQLFN NLGLCCFYAQ QYDMTLTSFE RALSLAENEE EAAD VWYNL GHVAVGIGDT NLAHQCFRLA LVNNNNHAEA YNNLAVLEMR KGHVEQARAL LQTASSLAPH MYEPHFNFAT ISDKI GDLQ RSYVAAQKSE AAFPDHVDTQ HLIKQLRQHF AML UniProtKB: Tetratricopeptide repeat protein 8 |
-Macromolecule #4: Protein PTHB1
| Macromolecule | Name: Protein PTHB1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99.383914 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSLFKARDWW STILGDKEEF DQGCLCLANV DNSGNGQDKI IVGSFMGYLR IFSPHPAKTG DGAQAEDLLL EVDLRDPVLQ VEVGKFVSG TEMLHLAVLH SRKLCVYSVS GTLGNVEHGN QCQMKLMYEH NLQRTACNMT YGSFGGVKGR DLICIQSMDG M LMVFEQES ...String: MSLFKARDWW STILGDKEEF DQGCLCLANV DNSGNGQDKI IVGSFMGYLR IFSPHPAKTG DGAQAEDLLL EVDLRDPVLQ VEVGKFVSG TEMLHLAVLH SRKLCVYSVS GTLGNVEHGN QCQMKLMYEH NLQRTACNMT YGSFGGVKGR DLICIQSMDG M LMVFEQES YAFGRFLPGF LLPGPLAYSS RTDSFLTVSS CQQVESYKYQ VLAFATDADK RQETEQQKLG SGKRLVVDWT LN IGEQALD ICIVSFNQSA SSVFVLGERN FFCLKDNGQI RFMKKLDWSP SCFLPYCSVS EGTINTLIGN HNNMLHIYQD VTL KWATQL PHIPVAVRVG CLHDLKGVIV TLSDDGHLQC SYLGTDPSLF QAPNVQSREL NYDELDVEMK ELQKIIKDVN KSQG VWPMT EREDDLNVSV VVSPNFDSVS QATDVEVGTD LVPSVTVKVT LQNRVILQKA KLSVYVQPPL ELTCDQFTFE FMTPD LTRT VSFSVYLKRS YTPSELEGNA VVSYSRPTDR NPDGIPRVIQ CKFRLPLKLI CLPGQPSKTA SHKITIDTNK SPVSLL SLF PGFASQSDDD QVNVMGFHFL GGARITVLAS KTSQRYRIQS EQFEDLWLIT NELILRLQEY FEKQGVKDFA CSFSGSI PL QEYFELIDHH FELRINGEKL EELLSERAVQ FRAIQRRLLA RFKDKTPAPL QHLDTLLDGT YKQVIALADA VEENQGNL F QSFTRLKSAT HLVILLIALW QKLSADQVAI LEAAFLPLQE DTQELGWEET VDAAISHLLK TCLSKSSKEQ ALNLNSQLN IPKDTSQLKK HITLLCDRLS KGGRLCLSTD AAAPQTMVMP GGCTTIPESD LEERSVEQDS TELFTNHRHL TAETPRPEVS PLQGVSE UniProtKB: Protein PTHB1 |
-Macromolecule #5: BBSome-interacting protein 1
| Macromolecule | Name: BBSome-interacting protein 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.430824 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASWSHPQFE KGSAGSAAGS GAGWSHPQFE KGAGLEVLFQ GPKRAEFMLK AAAKRPELSG KNTISNNSDM AEVKSMFREV LPKQGPLFV EDIMTMVLCK PKLLPLKSLT LEKLEKMHQA AQNTIRQQEM AEKDQRQITH UniProtKB: BBSome-interacting protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 286 K / Instrument: FEI VITROBOT MARK III Details: double blot with 2 minutes incubation after first sample application. | ||||||||
| Details | The sample was cross linked with 0.5% glutaraldehyde |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-50 / Number real images: 15266 / Average exposure time: 15.0 sec. / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 1.0 µm / Calibrated defocus min: 0.3 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)