+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-0541 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of a MAPK pathway complex | |||||||||
 マップデータ マップデータ | Structure of a MAPK pathway complex | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | TRANSFERASE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報epithelial cell proliferation involved in lung morphogenesis / positive regulation of endodermal cell differentiation / negative regulation of homotypic cell-cell adhesion / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / regulation of vascular associated smooth muscle contraction / positive regulation of axon regeneration / CD4-positive, alpha-beta T cell differentiation / mitogen-activated protein kinase kinase / CD4-positive or CD8-positive, alpha-beta T cell lineage commitment / negative regulation of synaptic vesicle exocytosis ...epithelial cell proliferation involved in lung morphogenesis / positive regulation of endodermal cell differentiation / negative regulation of homotypic cell-cell adhesion / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / regulation of vascular associated smooth muscle contraction / positive regulation of axon regeneration / CD4-positive, alpha-beta T cell differentiation / mitogen-activated protein kinase kinase / CD4-positive or CD8-positive, alpha-beta T cell lineage commitment / negative regulation of synaptic vesicle exocytosis / positive regulation of muscle contraction / Golgi inheritance / placenta blood vessel development / MAP-kinase scaffold activity / regulation of axon regeneration / cerebellar cortex formation / Signalling to p38 via RIT and RIN / labyrinthine layer development / melanosome transport / head morphogenesis / ARMS-mediated activation / type B pancreatic cell proliferation / endothelial cell apoptotic process / myeloid progenitor cell differentiation / Signaling by MAP2K mutants / SHOC2 M1731 mutant abolishes MRAS complex function / Gain-of-function MRAS complexes activate RAF signaling / negative regulation of fibroblast migration / positive regulation of D-glucose transmembrane transport / vesicle transport along microtubule / establishment of protein localization to membrane / positive regulation of axonogenesis / positive regulation of Ras protein signal transduction / regulation of Golgi inheritance / mitogen-activated protein kinase kinase kinase binding / central nervous system neuron differentiation / regulation of T cell differentiation / trachea formation / triglyceride homeostasis / Negative feedback regulation of MAPK pathway / regulation of early endosome to late endosome transport / regulation of stress-activated MAPK cascade / Frs2-mediated activation / stress fiber assembly / MAPK3 (ERK1) activation / ERBB2-ERBB3 signaling pathway / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / face development / endodermal cell differentiation / MAP kinase kinase activity / Bergmann glial cell differentiation / positive regulation of ATP biosynthetic process / thyroid gland development / Uptake and function of anthrax toxins / positive regulation of protein serine/threonine kinase activity / synaptic vesicle exocytosis / somatic stem cell population maintenance / positive regulation of peptidyl-serine phosphorylation / MAP kinase kinase kinase activity / protein kinase activator activity / negative regulation of endothelial cell apoptotic process / Schwann cell development / response to axon injury / postsynaptic modulation of chemical synaptic transmission / keratinocyte differentiation / positive regulation of stress fiber assembly / neuron projection morphogenesis / ERK1 and ERK2 cascade / myelination / positive regulation of substrate adhesion-dependent cell spreading / positive regulation of autophagy / protein serine/threonine/tyrosine kinase activity / dendrite cytoplasm / substrate adhesion-dependent cell spreading / insulin-like growth factor receptor signaling pathway / cellular response to calcium ion / response to glucocorticoid / thymus development / animal organ morphogenesis / MAP3K8 (TPL2)-dependent MAPK1/3 activation / protein serine/threonine kinase activator activity / Signal transduction by L1 / cell motility / positive regulation of transcription elongation by RNA polymerase II / RAF activation / Spry regulation of FGF signaling / Signaling by high-kinase activity BRAF mutants / MAP2K and MAPK activation / visual learning / small GTPase binding / cellular response to xenobiotic stimulus / centriolar satellite / epidermal growth factor receptor signaling pathway / long-term synaptic potentiation / neuron differentiation / chemotaxis / Negative regulation of MAPK pathway / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) / Homo sapiens (ヒト) /  Spodoptera exigua (シロイチモジヨトウ) Spodoptera exigua (シロイチモジヨトウ) | |||||||||
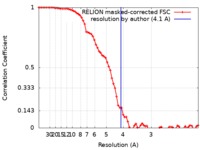
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.1 Å | |||||||||
 データ登録者 データ登録者 | Park E / Rawson S | |||||||||
| 資金援助 |  米国, 2件 米国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2019 ジャーナル: Nature / 年: 2019タイトル: Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. 著者: Eunyoung Park / Shaun Rawson / Kunhua Li / Byeong-Won Kim / Scott B Ficarro / Gonzalo Gonzalez-Del Pino / Humayun Sharif / Jarrod A Marto / Hyesung Jeon / Michael J Eck /  要旨: RAF family kinases are RAS-activated switches that initiate signalling through the MAP kinase cascade to control cellular proliferation, differentiation and survival. RAF activity is tightly ...RAF family kinases are RAS-activated switches that initiate signalling through the MAP kinase cascade to control cellular proliferation, differentiation and survival. RAF activity is tightly regulated and inappropriate activation is a frequent cause of cancer; however, the structural basis for RAF regulation is poorly understood at present. Here we use cryo-electron microscopy to determine autoinhibited and active-state structures of full-length BRAF in complexes with MEK1 and a 14-3-3 dimer. The reconstruction reveals an inactive BRAF-MEK1 complex restrained in a cradle formed by the 14-3-3 dimer, which binds the phosphorylated S365 and S729 sites that flank the BRAF kinase domain. The BRAF cysteine-rich domain occupies a central position that stabilizes this assembly, but the adjacent RAS-binding domain is poorly ordered and peripheral. The 14-3-3 cradle maintains autoinhibition by sequestering the membrane-binding cysteine-rich domain and blocking dimerization of the BRAF kinase domain. In the active state, these inhibitory interactions are released and a single 14-3-3 dimer rearranges to bridge the C-terminal pS729 binding sites of two BRAFs, which drives the formation of an active, back-to-back BRAF dimer. Our structural snapshots provide a foundation for understanding normal RAF regulation and its mutational disruption in cancer and developmental syndromes. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_0541.map.gz emd_0541.map.gz | 21.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-0541-v30.xml emd-0541-v30.xml emd-0541.xml emd-0541.xml | 20.9 KB 20.9 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_0541_fsc.xml emd_0541_fsc.xml | 7.2 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_0541.png emd_0541.png | 188 KB | ||
| Filedesc metadata |  emd-0541.cif.gz emd-0541.cif.gz | 7.8 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0541 http://ftp.pdbj.org/pub/emdb/structures/EMD-0541 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0541 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0541 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_0541_validation.pdf.gz emd_0541_validation.pdf.gz | 439.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_0541_full_validation.pdf.gz emd_0541_full_validation.pdf.gz | 438.9 KB | 表示 | |
| XML形式データ |  emd_0541_validation.xml.gz emd_0541_validation.xml.gz | 9.6 KB | 表示 | |
| CIF形式データ |  emd_0541_validation.cif.gz emd_0541_validation.cif.gz | 12.4 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0541 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0541 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0541 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0541 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_0541.map.gz / 形式: CCP4 / 大きさ: 30.5 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_0541.map.gz / 形式: CCP4 / 大きさ: 30.5 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Structure of a MAPK pathway complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Ternary complex of BRAF/MEK1/14-3-3 with MEK inhibitor
| 全体 | 名称: Ternary complex of BRAF/MEK1/14-3-3 with MEK inhibitor |
|---|---|
| 要素 |
|
-超分子 #1: Ternary complex of BRAF/MEK1/14-3-3 with MEK inhibitor
| 超分子 | 名称: Ternary complex of BRAF/MEK1/14-3-3 with MEK inhibitor タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#3 詳細: 14-3-3 is heterodimer of Insect epsilon and zeta. model was generated by Insect zeta sequence as a homo-dimer. |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 190 KDa |
-分子 #1: Serine/threonine-protein kinase B-raf
| 分子 | 名称: Serine/threonine-protein kinase B-raf / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO / EC番号: non-specific serine/threonine protein kinase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 89.402789 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSYYHHHHHH HHDIPTTENL YFQGAMDMAA LSGGGGGGAE PGQALFNGDM EPEAGAGAGA AASSAADPAI PEEVWNIKQM IKLTQEHIE ALLDKFGGEH NPPSIYLEAY EEYTSKLDAL QQREQQLLES LGNGTDFSVS SSASMDTVTS SSSSSLSVLP S SLSVFQNP ...文字列: MSYYHHHHHH HHDIPTTENL YFQGAMDMAA LSGGGGGGAE PGQALFNGDM EPEAGAGAGA AASSAADPAI PEEVWNIKQM IKLTQEHIE ALLDKFGGEH NPPSIYLEAY EEYTSKLDAL QQREQQLLES LGNGTDFSVS SSASMDTVTS SSSSSLSVLP S SLSVFQNP TDVARSNPKS PQKPIVRVFL PNKQRTVVPA RCGVTVRDSL KKALMMRGLI PECCAVYRIQ DGEKKPIGWD TD ISWLTGE ELHVEVLENV PLTTHNFVRK TFFTLAFCDF CRKLLFQGFR CQTCGYKFHQ RCSTEVPLMC VNYDQLDLLF VSK FFEHHP IPQEEASLAE TALTSGSSPS APASDSIGPQ ILTSPSPSKS IPIPQPFRPA DEDHRNQFGQ RDRSS(SEP)APNV HINTIEPVN IDDLIRDQGF RGDGGSTTGL SATPPASLPG SLTNVKALQK SPGPQRERKS SSSSEDRNRM KTLGRRDSSD D WEIPDGQI TVGQRIGSGS FGTVYKGKWH GDVAVKMLNV TAPTPQQLQA FKNEVGVLRK TRHVNILLFM GYSTKPQLAI VT QWCEGSS LYHHLHIIET KFEMIKLIDI ARQTAQGMDY LHAKSIIHRD LKSNNIFLHE DLTVKIGDFG LATVKSRWSG SHQ FEQLSG SILWMAPEVI RMQDKNPYSF QSDVYAFGIV LYELMTGQLP YSNINNRDQI IFMVGRGYLS PDLSKVRSNC PKAM KRLMA ECLKKKRDER PLFPQILASI ELLARSLPKI HRSA(SEP)EPSLN RAGFQTEDFS LYACASPKTP IQAGGYGAFP V HGTSAWSH PQFEK UniProtKB: Serine/threonine-protein kinase B-raf |
-分子 #2: Dual specificity mitogen-activated protein kinase kinase 1
| 分子 | 名称: Dual specificity mitogen-activated protein kinase kinase 1 タイプ: protein_or_peptide / ID: 2 / 詳細: GDC-0623 / コピー数: 1 / 光学異性体: LEVO / EC番号: mitogen-activated protein kinase kinase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 45.934543 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MGSSHHHHHH SAVDENLYFQ GGMPKKKPTP IQLNPAPDGS AVNGTSSAET NLEALQKKLE ELELDEQQRK RLEAFLTQKQ KVGELKDDD FEKISELGAG NGGVVFKVSH KPSGLVMARK LIHLEIKPAI RNQIIRELQV LHECNSPYIV GFYGAFYSDG E ISICMEHM ...文字列: MGSSHHHHHH SAVDENLYFQ GGMPKKKPTP IQLNPAPDGS AVNGTSSAET NLEALQKKLE ELELDEQQRK RLEAFLTQKQ KVGELKDDD FEKISELGAG NGGVVFKVSH KPSGLVMARK LIHLEIKPAI RNQIIRELQV LHECNSPYIV GFYGAFYSDG E ISICMEHM DGGSLDQVLK KAGRIPEQIL GKVSIAVIKG LTYLREKHKI MHRDVKPSNI LVNSRGEIKL CDFGVSGQLI DA MANAFVG TRSYMSPERL QGTHYSVQSD IWSMGLSLVE MAVGRYPIPP PDAKELELMF GCQVEGDAAE TPPRPRTPGR PLS SYGMDS RPPMAIFELL DYIVNEPPPK LPSGVFSLEF QDFVNKCLIK NPAERADLKQ LMVHAFIKRS DAEEVDFAGW LCST IGLNQ PSTPTHAAGV UniProtKB: Dual specificity mitogen-activated protein kinase kinase 1 |
-分子 #3: 14-3-3 protein zeta
| 分子 | 名称: 14-3-3 protein zeta / タイプ: protein_or_peptide / ID: 3 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Spodoptera exigua (シロイチモジヨトウ) Spodoptera exigua (シロイチモジヨトウ) |
| 分子量 | 理論値: 28.108514 KDa |
| 配列 | 文字列: MSVDKEELVQ RAKLAEQAER YDDMAAAMKE VTETGVELSN EERNLLSVAY KNVVGARRSS WRVISSIEQK TEGSERKQQM AKEYRVKVE KELREICYDV LGLLDKHLIP KASNPESKVF YLKMKGDYYR YLAEVATGET RNSVVEDSQK AYQDAFEISK A KMQPTHPI ...文字列: MSVDKEELVQ RAKLAEQAER YDDMAAAMKE VTETGVELSN EERNLLSVAY KNVVGARRSS WRVISSIEQK TEGSERKQQM AKEYRVKVE KELREICYDV LGLLDKHLIP KASNPESKVF YLKMKGDYYR YLAEVATGET RNSVVEDSQK AYQDAFEISK A KMQPTHPI RLGLALNFSV FYYEILNSPD KACQLAKQAF DDAIAELDTL NEDSYKDSTL IMQLLRDNLT LWTSDTQGDG DE PAEGGDN UniProtKB: 14-3-3 protein zeta |
-分子 #4: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| 分子 | 名称: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / タイプ: ligand / ID: 4 / コピー数: 1 / 式: AGS |
|---|---|
| 分子量 | 理論値: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-分子 #5: ZINC ION
| 分子 | 名称: ZINC ION / タイプ: ligand / ID: 5 / コピー数: 2 / 式: ZN |
|---|---|
| 分子量 | 理論値: 65.409 Da |
-分子 #6: ADENOSINE-5'-DIPHOSPHATE
| 分子 | 名称: ADENOSINE-5'-DIPHOSPHATE / タイプ: ligand / ID: 6 / コピー数: 1 / 式: ADP |
|---|---|
| 分子量 | 理論値: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-分子 #7: MAGNESIUM ION
| 分子 | 名称: MAGNESIUM ION / タイプ: ligand / ID: 7 / コピー数: 1 / 式: MG |
|---|---|
| 分子量 | 理論値: 24.305 Da |
-分子 #8: 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a...
| 分子 | 名称: 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide タイプ: ligand / ID: 8 / コピー数: 1 / 式: LCJ |
|---|---|
| 分子量 | 理論値: 456.21 Da |
| Chemical component information | 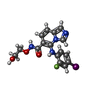 ChemComp-LCJ: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.05 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 |
| グリッド | 詳細: unspecified |
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 照射モード: OTHER / 撮影モード: OTHER / Cs: 2.7 mm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)