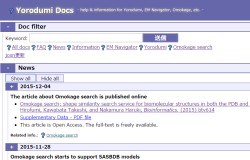
-Search & filter documents
 FAQ,
FAQ,  News,
News,  Information,
Information,  EM Navigator,
EM Navigator,  Yorodumi,
Yorodumi,  Omokage search,
Omokage search,  Services,
Services,  All docs
All docs
-Information (43)
+EMDB map data format
3D density map data in CCP4 format
Related info.: EMDB /
EMDB /  Q: What is the format for the maps? /
Q: What is the format for the maps? /  Q: Which software is suitable to view EMDB maps? /
Q: Which software is suitable to view EMDB maps? /  Q: Is an EM map electron-density map?
Q: Is an EM map electron-density map?
External links: EMDB Map Distribution Format Description /
EMDB Map Distribution Format Description /  Format technical details /
Format technical details /  CCP4 - Wikipedia
CCP4 - Wikipedia
+EMDB header
Meta information in XML format
Data file in XML format containing information associated with the map of EMDB entries, experimental information, detailed sample information, etc.
Related info.: EMDB /
EMDB /  Feb 9, 2022. New format data for meta-information of EMDB entries
Feb 9, 2022. New format data for meta-information of EMDB entries
External links: EMDB Schema v3 /
EMDB Schema v3 /  EMDB Schema v1.9 /
EMDB Schema v1.9 /  Documentation directory
Documentation directory
+Mask map
Map data of mask pattern, sub-region, segmentation, etc
Related info.: EMDB
EMDB
+FSC data file
Fourier shell correlation data in XML format for resolution estimation
Related info.: EMDB
EMDB
External links: Fourier shell correlation - Wikipedia /
Fourier shell correlation - Wikipedia /  FSC-schema
FSC-schema
+EMDB validaton report
Validaton report of EMDB entry by wwPDB and EMDB
Related info.: EMDB /
EMDB /  wwPDB validaton report
wwPDB validaton report
External links: User guide to the EmDataBank map validation reports
User guide to the EmDataBank map validation reports
+PDBx/mmCIF format
Officail data format for PDB entries, in CIF text format, having atomic coordinates and meta information
Related info.: PDB /
PDB /  Q: What is difference in PDB coordinate formats?
Q: What is difference in PDB coordinate formats?
External links: PDBx/mmCIF Dictionary Resources /
PDBx/mmCIF Dictionary Resources /  Crystallographic Information File
Crystallographic Information File
+PDB format
Legacy officail data format for PDB entries, contains atomic coordinates
Related info.: PDB /
PDB /  Q: What is difference in PDB coordinate formats?
Q: What is difference in PDB coordinate formats?
External links: Atomic Coordinate Entry Format Version 3.3
Atomic Coordinate Entry Format Version 3.3
+PDBx/mmJSON format
JSON representation of the PDBx/mmCIF data developed by PDBj
Related info.: PDB /
PDB /  Q: What is difference in PDB coordinate formats?
Q: What is difference in PDB coordinate formats?
External links: mmJSON - PDBj help
mmJSON - PDBj help
+wwPDB validaton report
Validaton report of PDB entry by wwPDB
Related info.: PDB /
PDB /  EMDB validaton report
EMDB validaton report
External links: wwPDB: Validation Reports /
wwPDB: Validation Reports /  wwPDB: validation report FAQs
wwPDB: validation report FAQs
+EM Navigator
3D electron microscopy data browser

URL: https://pdbj.org/emnavi/.
- EM Navigator is a browser for 3D electron microscopy (3D-EM) data of biological molecules and assemblies.
- It provides EMDB-PDB cross-search, statistical information, and links to similar structures.
- You can easily check the latest EM structural data and structural papers.
Related info.: EMDB /
EMDB /  PDB /
PDB /  PDBj /
PDBj /  Q: What are the data sources of EM Navigator? /
Q: What are the data sources of EM Navigator? /  EMN Search /
EMN Search /  EMN Papers /
EMN Papers /  EMN Gallery /
EMN Gallery /  EMN Statistics /
EMN Statistics /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
+EMN Search
3DEM data search

URL: https://pdbj.org/emnavi/esearch.php
Advanced data search for EMDB and EM data in PDB widh various search and display options
Related info.: EMDB /
EMDB /  PDB /
PDB /  EM Navigator /
EM Navigator /  Q: What are the data sources of EM Navigator? /
Q: What are the data sources of EM Navigator? /  Yorodumi Search /
Yorodumi Search /  Jul 5, 2019. Downlodablable text data
Jul 5, 2019. Downlodablable text data
+EMN Papers
Database of articles cited by 3DEM data entries

URL: https://pdbj.org/emnavi/pap.php?em=1
- Database of articles cited by 3DEM data entries in EMDB and PDB
- Using PubMed data
Related info.: EMDB /
EMDB /  PDB /
PDB /  Q: What are the data sources of EM Navigator? /
Q: What are the data sources of EM Navigator? /  EM Navigator /
EM Navigator /  Yorodumi Papers /
Yorodumi Papers /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
+EMN Gallery
Image gallery of 3DEM data

URL: https://pdbj.org/emnavi/gallery.php
Categorization is done by EM Navigator manager manually. It is not strict.
Related info.: EM Navigator
EM Navigator
+EMN Statistics
Statistics of 3DEM data in table and graph styles

URL: https://pdbj.org/emnavi/stat.php
- The table can be sorted. Click the column header to be sorted. (second click to reverse, [Shift]+click for multi-column sort)
- To show the bar graph in table mode, point the column/row header by mouse courser.
- To search the correspoinding data, click the cell of value.
- Examples:
Related info.: Q: What are the data sources of EM Navigator? /
Q: What are the data sources of EM Navigator? /  EM Navigator /
EM Navigator /  Mar 30, 2013. New page of EM Navigator, EMN Statistics
Mar 30, 2013. New page of EM Navigator, EMN Statistics
+Yorodumi
Thousand views of thousand structures
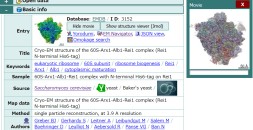
URL: https://pdbj.org/emnavi/quick.php
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
+Yorodumi Search
Cross-search of EMDB, PDB, SASBDB, etc.
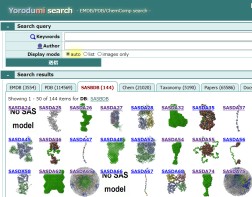
URL: https://pdbj.org/emnavi/ysearch.php
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  EMN Search /
EMN Search /  Yorodumi /
Yorodumi /  Experimental methods, equipments, and software data /
Experimental methods, equipments, and software data /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
+Yorodumi Papers
Database of articles cited by EMDB/PDB/SASBDB data
URL: https://pdbj.org/emnavi/pap.php
- Database of articles cited by EMDB, PDB, and SASBDB entries
- Using PubMed data
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Yorodumi /
Yorodumi /  EMN Papers /
EMN Papers /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
+Yorodumi Speices
Taxonomy data in EMDB/PDB/SASBDB
URL: https://pdbj.org/emnavi/taxo.php
Taxonomy database of sample sources of data in EMDB/PDB/SASBDB
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Omokage search
Search structure by SHAPE

URL: https://pdbj.org/omokage/
- "Omokage search" is a shape similarity search service for 3D structures of macromolecules. By comparing global shapes, and ignoring details, similar-shaped structures are searched.
- The search is performed ageinst >200,000 structure data, which consists of EMDB map data, PDB coordinates (deposited units (asymmetric units, usually), PDB biological units, and SASBDB mdoels).
- For the search query, you can use either a data in the PDB/EMDB/SASBDB or your original model.
- Supported formats are PDB (atomic model, SAXS bead model, etc.) and CCP4/MRC map (3D density map).
- Shape comparison is performed by iDR profile method, which uses 1D-distance profiles of super-simplified models generated by vector quantizaion in Situs package.
Related info.: Comparison of 3 databanks /
Comparison of 3 databanks /  Sep 22, 2014. New service: Omokage search /
Sep 22, 2014. New service: Omokage search /  gmfit /
gmfit /  Re-ranking by gmfit
Re-ranking by gmfit
+Yorodumi Docs
+Covid-19 info
Page for Covid-19 featured contents

URL: https://pdbj.org/emnavi/covid19.php
Related info.: Comparison of 3 databanks /
Comparison of 3 databanks /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
+F&H Search
Function & Homology similarity search
URL: https://pdbj.org/emnavi/fh-search.php
Based on the similarity of Function & Homolog items related to the structure data, similar data are searched from structure databases.
Related info.: Function and homology information
Function and homology information
+Movie viewer
Visualization of 3DEM structure data by movies
- Horizontal- and vertical- rotation motions and "slicing" motion are recorded in the movies.
- As the movie frame can be controlled by mouse/touch position, it can be operated interactively as a molecular viewer.
- According to the mouse/finger position and orientation of the motion on the movie panel, the model rotates on horizontal or vertical direction.
- By horizontal mouse/finger movement on the top part of the movie, the slicing motion is shown.
Related info.: Q: How do you make the movies in EM Navigator? /
Q: How do you make the movies in EM Navigator? /  Q: Can I use the movies and their snapshots in the EM Navigator for papers or presentations? /
Q: Can I use the movies and their snapshots in the EM Navigator for papers or presentations? /  SurfView /
SurfView /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
+Jmol/JSmol
An open-source viewer for chemical structures in 3D
- Mouse/touch operation
X-Y rotation X-Y move Zoom in/out Mouse left drag [Ctrl] + right drag center drag (vertical) / wheel Touch single finger double finger pinch - Jmol menu: [Ctrl] + click / right click
Related info.: Molmil /
Molmil /  Yorodumi
Yorodumi
External links: Jmol: an open-source Java viewer for chemical structures in 3D
Jmol: an open-source Java viewer for chemical structures in 3D
+Molmil
WebGL based molecular viewer
- Molmil runs on Web browser on a smartphone or tablet device as well as PC. (Molmil is based on WebGL and Javascript).
- Mouse/touch operation
X-Y rotation X-Y move Zoom in/out Mouse left drag center drag / shift + left drag right drag / wheel Touch single finger double finger pinch - Function menu: top left buttons on viewer screen
Related info.: Jmol/JSmol /
Jmol/JSmol /  SurfView /
SurfView /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
External links: Documentation of Molmil in PDBj site /
Documentation of Molmil in PDBj site /  Molmil page in GitHub
Molmil page in GitHub
+SurfView
EMDB map surface models viewer on Web browsers

- SurfView run on Web browser on a smartphone or tablet device as well as PC.
- SurfView is based on a WebGL and three.js, Javascript 3D library.
- The surface models shwon in this viewer are modified for simplification and data size reduction. To get views of full-resolution data, see movies instead.
- Mouse/touch operation%table-1%
Related info.: Movie viewer /
Movie viewer /  Molmil /
Molmil /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
+Comparison of 3 databanks
Yorodumi and Omokage can cross-search these DBs
| Method | Main data | Str. data format | meta data format | |
|---|---|---|---|---|
| PDB | various | atomic model | PDBx/mmCIF, etc. | PDBx/mmCIF, etc. |
| EMDB | 3DEM | 3D map | CCP4 map | EMDB XML |
| SASBDB | Small Angle Scattering (SAS) | SAS profile (+/- 3D models) | PDB + sasCIF | ASCII + sasCIF |
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Covid-19 info /
Covid-19 info /  ID/Accession-code notation in Yorodumi/EM Navigator /
ID/Accession-code notation in Yorodumi/EM Navigator /  Experimental methods, equipments, and software data
Experimental methods, equipments, and software data
+ID/Accession-code notation in Yorodumi/EM Navigator
| Database | pattern | e.g. | note |
|---|---|---|---|
| EMDB | EMDB-**** | EMDB-1001 | prefix EMD- omitted |
| PDB | PDB-**** | PDB-1a00 | |
| SASBDB | _ | SASDA24 | as it is |
Related info.: Comparison of 3 databanks /
Comparison of 3 databanks /  Q: What is EMD? /
Q: What is EMD? /  Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
+EMDB
Electron Microscopy Data Bank, Databank for 3DEM

URL: https://www.ebi.ac.uk/emdb/
Related info.: 3D electron microscopy (3DEM) /
3D electron microscopy (3DEM) /  PDB /
PDB /  Comparison of 3 databanks
Comparison of 3 databanks
External links: The Electron Microscopy Data Bank - EBI /
The Electron Microscopy Data Bank - EBI /  EMDataResource
EMDataResource
+PDB
Protein Data Bank - The single repository of information about the 3D structures of proteins, nucleic acids, and complex assemblies

URL: https://wwPDB.org/
Related info.: PDBj /
PDBj /  EMDB
EMDB
+PDBj
Protein Data Bank Japan

URL: http://pdbj.org/
A member of wwPDB
Related info.: PDB /
PDB /  Information from PDBMLplus
Information from PDBMLplus
External links: Protein Data Bank Japan at IPR, Osaka-univ. /
Protein Data Bank Japan at IPR, Osaka-univ. /  PDBj@Facebook /
PDBj@Facebook /  PDBj@Twitter
PDBj@Twitter
+SASBDB
Small Angle Scattering Biological Data Bank - Curated repository for small angle scattering data and models

- Develop & manage: BioSAXS group in EMBL
- Since 2014
Related info.: Comparison of 3 databanks
Comparison of 3 databanks
+gmfit
A program for fitting subunits into density map of complex using GMM (Gaussian Mixture Model)

- Developed by Takeshi Kawabata in IPR, Osaka-univ.
- Used from Omokage search.
Related info.: Omokage search /
Omokage search /  Dec 4, 2015. The article about Omokage search is published online /
Dec 4, 2015. The article about Omokage search is published online /  Re-ranking by gmfit
Re-ranking by gmfit
External links: Pairwise gmfit
Pairwise gmfit
+3D electron microscopy (3DEM)
A generic term of electron microscopic analyses to obtain 3D structures
- Analyses such as electron tomography, single particle analysis, and electron diffraction are included.
Aggregation states Methods generally used "individual structure" electron tomography "single particle" single particle analysis "icosahedral" single particle analysis "helical" helical reconstruction / single particle analysis "2D/3D-crystal" electron diffraction / Fourier filtering - Characteristics compared to X-ray crystallography and NMR:
Advantage Wider applicability of sample (not require high-purity or high-concentration smaple, and useful for more huge, complex, flexible, and less uniform sample) Disadvantage (previous) Lower resolution (hard to get atomic-level resolution data) Disadvantage (new) High-performance electron microscopes and their maintenance costs are quite expensive
Related info.: EMDB /
EMDB /  electron cryo microscopy (cryoEM)
electron cryo microscopy (cryoEM)
+electron cryo microscopy (cryoEM)
Electron microscopy where the sample is studied at cryogenic temperatures
The specimen is cooled at 4~100K (-269~-170℃) to keep it in hydrated state in the highly vaqumed environment, and to reduce radiation dammage by electron beam.
Related info.: 3D electron microscopy (3DEM) /
3D electron microscopy (3DEM) /  Q: Is 3DEM same as electron cryo microscopy (cryoEM)?
Q: Is 3DEM same as electron cryo microscopy (cryoEM)?
+"Movies out of date"
Movies with this annotation may not be up-to-date.
EMDB map data are sometimes remediated. The process of making movies in the EM Navigator are not fully automated, and it is very hard to remake the all to catch up. So, some movies and movie parameters are based on the older data, and may be improper for new map data.
Related info.: Q: How do you make the movies in EM Navigator?
Q: How do you make the movies in EM Navigator?
+Carbohydrate representation
Polysaccaride/carbohydrate data representation in PDB
- In July 2020, representation for polysaccaride/carbohydrate information in PDB is improved.
- The polysaccaride figures are generated by GlycanBuilder2.
- See following external links for details.
External links: Implementation of GlycanBuilder to draw a wide variety of ambiguous glycans - ScienceDirect /
Implementation of GlycanBuilder to draw a wide variety of ambiguous glycans - ScienceDirect /  GlycanBuilder2 - RINGS (Resource For INformatics Of Glycomes at Soka) /
GlycanBuilder2 - RINGS (Resource For INformatics Of Glycomes at Soka) /  Carbohydrate Remediation - wwPDB documentation /
Carbohydrate Remediation - wwPDB documentation /  Coming July 29: Improved Carbohydrate Data at the PDB - wwPDB news
Coming July 29: Improved Carbohydrate Data at the PDB - wwPDB news
+Experimental methods, equipments, and software data
Database of experimental methods of EMDB, PDB and SABDB.
URL: https://pdbj.org/emnavi/ysearch.php?&act_tab=met
Related info.: Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search
Yorodumi Search
+Function and homology information
Molecular function, domain, and homology information from related databases.
- To help to understand the molecular function and to find the related structure data, Yorodumi and Yorodumi Search display and utilize the related database information about function and homology.
- In addition to the information of EC, EMBL, GeneBank, GO, InterPro, UniProt, etc. stored in the EMDB header XML, PDBx/mmCIF and sasCIF original data, information of Pfam, PROSITE, Reactome, UniProkKB, etc. are collected via PDBMLplus, EMDB-PDB fitting data, and/or UniProt.
Category Name of database or definition Function% Gene ontology, Enzyme Commission number, Reactome, etc. Domain/homology CATH, InterPro, SMART, etc. Component UniProt, GenBank, PDB chemical component, etc.
Related info.: F&H Search /
F&H Search /  Information from PDBMLplus /
Information from PDBMLplus /  Yorodumi Search /
Yorodumi Search /  Yorodumi /
Yorodumi /  F&H Search
F&H Search
+Re-ranking by gmfit
The similarity ranking by Omokage search can be re-ordered according to correlation coefficient by gmfit, expecting incorporation of the (potential) benefits of two methods with following properties.
| Name | Method | Speed | Potential accuracy |
|---|---|---|---|
| Omokage search | iDR profile comparison | ++ sub-msec / 1 comparison | - 1D profile comparison |
| gmfit | Gaussian mixture model fitting | + sub-sec / 1 comparison | + comparison in 3D space |
Related info.: gmfit /
gmfit /  Omokage search
Omokage search
+Changes in new EM Navigator and Yorodumi
Changes in new versions of EM Navigator and Yorodumi. (Sep. 2016)
- Changes:
- New user interface unified in EM Navigator, Yorodumi & Omokage search, supporting mobile devices as well as PCs.
- New Yorodumi replace the individual data page ("Detail page" of EM Navigator in the legacy system). It is unified browser for EMDB, PDB, and SASBDB entries integrated with EM Navigator and Omokage search.
- The viewers (structure/movie viewers) appear in pop-up windows. On a PC, multiple viewer windows can be opened. On mobile devices, the viewers support touch operation.
- New features:
- Molmil, molecular structure viewer, and SurfView, surface model viewer for EMDB map data, are available to view the 3D structures. Both viewers support mobile device use.
- Yorodumi support SASBDB entries.
- Some new pages such as Yorodumi Papers, citation database of structure data entries and Yorodumi Search, cross search by keywords
Related info.: EM Navigator /
EM Navigator /  Yorodumi /
Yorodumi /  SurfView /
SurfView /  Molmil /
Molmil /  Movie viewer /
Movie viewer /  EMN Papers /
EMN Papers /  Yorodumi Papers /
Yorodumi Papers /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi
Aug 31, 2016. New EM Navigator & Yorodumi
+Information from PDBMLplus
PDBMLplus is the XML format file including additional information relating to individual proteins. Currently, PDB files are lacking a detailed description of function, experimental conditions, and the like. And then such information was included to extended XML database, named PDBMLplus.
Related info.: PDBj /
PDBj /  Function and homology information
Function and homology information
External links: PDBMLplus - PDBj helip /
PDBMLplus - PDBj helip /  About Functional Details page - PDBJ help
About Functional Details page - PDBJ help
+Yorodumi annotation
Annotation by Yorodumi/EM Navigator manager
Related info.: Q: Who make these documents? Who develop EM Navigator, Yorodumi, Omokage search, etc?
Q: Who make these documents? Who develop EM Navigator, Yorodumi, Omokage search, etc?
 Movie
Movie Controller
Controller Structure viewers
Structure viewers