7SHK
 
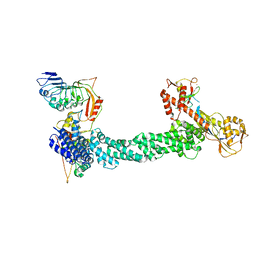 | | Structure of Xenopus laevis CRL2Lrr1 (State 1) | | Descriptor: | CULLIN_2 domain-containing protein, Elongin-C, Lrr1, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2021-10-09 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Nucleic Acids Res., 49, 2021
|
|
7SHL
 
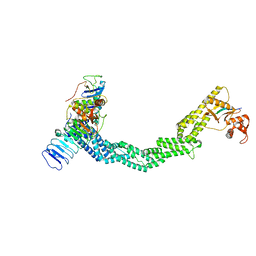 | | Structure of Xenopus laevis CRL2Lrr1 (State 2) | | Descriptor: | CULLIN_2 domain-containing protein, Elongin-C, Lrr1, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2021-10-09 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Nucleic Acids Res., 49, 2021
|
|
4E5Y
 
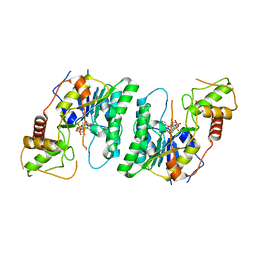 | |
3H3B
 
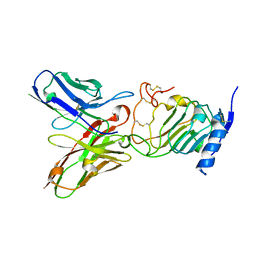 | | Crystal structure of the single-chain Fv (scFv) fragment of an anti-ErbB2 antibody chA21 in complex with residues 1-192 of ErbB2 extracellular domain | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, anti-ErbB2 antibody chA21 | | Authors: | Zhou, H, Liu, Y, Niu, L, Zhu, J, Teng, M. | | Deposit date: | 2009-04-16 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights into the Down-regulation of Overexpressed p185her2/neu Protein of Transformed Cells by the Antibody chA21.
J.Biol.Chem., 286, 2011
|
|
2HGF
 
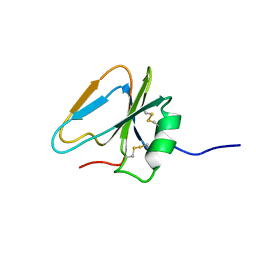 | | HAIRPIN LOOP CONTAINING DOMAIN OF HEPATOCYTE GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEPATOCYTE GROWTH FACTOR | | Authors: | Zhou, H, Mazzulla, M.J, Kaufman, J.D, Stahl, S.J, Wingfield, P.T, Rubin, J.S, Bottaro, D.P, Byrd, R.A. | | Deposit date: | 1997-12-18 | | Release date: | 1998-06-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site.
Structure, 6, 1998
|
|
4XEM
 
 | |
1T2M
 
 | | Solution Structure Of The Pdz Domain Of AF-6 | | Descriptor: | AF-6 protein | | Authors: | Zhou, H, Wu, J.H, Xu, Y.Q, Huang, A.D, Shi, Y.Y. | | Deposit date: | 2004-04-22 | | Release date: | 2005-02-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of AF-6 PDZ Domain and Its Interaction with the C-terminal Peptides from Neurexin and Bcr
J.Biol.Chem., 280, 2005
|
|
4HVC
 
 | | Crystal structure of human prolyl-tRNA synthetase in complex with halofuginone and ATP analogue | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Bifunctional glutamate/proline--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Zhou, H, Sun, L, Yang, X.L, Schimmel, P. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase.
Nature, 494, 2012
|
|
3Q19
 
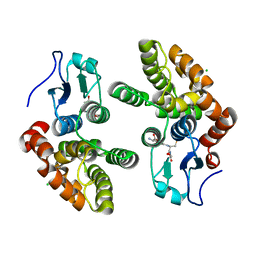 | | Human Glutathione Transferase O2 | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase omega-2 | | Authors: | Zhou, H, Board, P.G, Oakley, A.J. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
3PSM
 
 | |
3Q18
 
 | | Human Glutathione Transferase O2 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhou, H, Board, P.G, Oakley, A.J. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
3QAG
 
 | | Human Glutathione Transferase O2 with glutathione -new crystal form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Zhou, H, Board, P.G, Oakley, A.J. | | Deposit date: | 2011-01-11 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
6A68
 
 | | the crystal structure of rat calcium-dependent activator protein for secretion (CAPS) DAMH domain | | Descriptor: | Calcium-dependent secretion activator 1, POTASSIUM ION | | Authors: | Zhou, H, Wei, Z.Q, Yao, D.Q, Zhang, R.G, Ma, C. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural and Functional Analysis of the CAPS SNARE-Binding Domain Required for SNARE Complex Formation and Exocytosis.
Cell Rep, 26, 2019
|
|
6J11
 
 | | MERS-CoV spike N-terminal domain and 7D10 scFv complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-terminal domain of Spike glycoprotein, ... | | Authors: | Zhou, H, Zhang, S, Zhang, S, Tang, W, Wang, X. | | Deposit date: | 2018-12-27 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein.
Nat Commun, 10, 2019
|
|
6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ4
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ2
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ1
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6LAW
 
 | |
6LAV
 
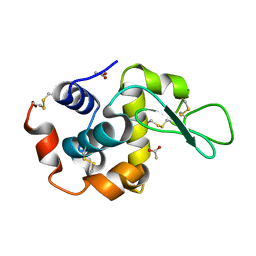 | |
5WXB
 
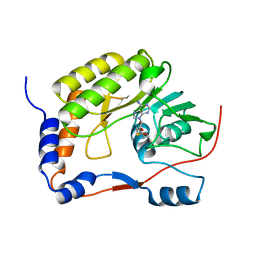 | |
5HXW
 
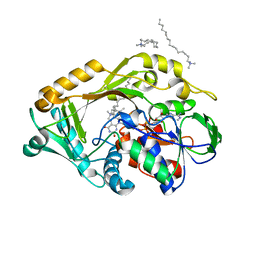 | | L-amino acid deaminase from Proteus vulgaris | | Descriptor: | CETYL-TRIMETHYL-AMMONIUM, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid deaminase | | Authors: | Zhou, H, Ju, Y, Niu, L, Teng, M. | | Deposit date: | 2016-01-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of a membrane-bound l-amino acid deaminase from Proteus vulgaris
J.Struct.Biol., 195, 2016
|
|
5I39
 
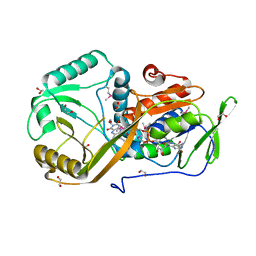 | | High resolution structure of L-amino acid deaminase from Proteus vulgaris with the deletion of the specific insertion sequence | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid deaminase | | Authors: | Zhou, H, Ju, Y, Niu, L, Teng, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a membrane-bound l-amino acid deaminase from Proteus vulgaris
J.Struct.Biol., 195, 2016
|
|
5V58
 
 | |
5V59
 
 | |
