1BG0
 
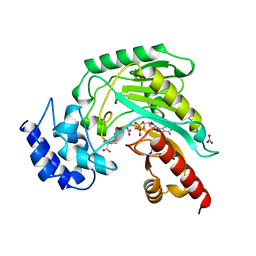 | | TRANSITION STATE STRUCTURE OF ARGININE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE KINASE, D-ARGININE, ... | | Authors: | Zhou, G, Somasundaram, T, Blanc, E, Parthasarathy, G, Ellington, W.R, Chapman, M.S. | | Deposit date: | 1998-06-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1FAV
 
 | | THE STRUCTURE OF AN HIV-1 SPECIFIC CELL ENTRY INHIBITOR IN COMPLEX WITH THE HIV-1 GP41 TRIMERIC CORE | | Descriptor: | HIV-1 ENVELOPE PROTEIN CHIMERA, PROTEIN (TRANSMEMBRANE GLYCOPROTEIN) | | Authors: | Zhou, G, Ferrer, M, Chopra, R, Strassmaier, T, Weissenhorn, W, Skehel, J.J, Oprian, D, Schreiber, S.L, Harrison, S.C, Wiley, D.C. | | Deposit date: | 2000-07-13 | | Release date: | 2000-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of an HIV-1 specific cell entry inhibitor in complex with the HIV-1 gp41 trimeric core.
Bioorg.Med.Chem., 8, 2000
|
|
1ZEO
 
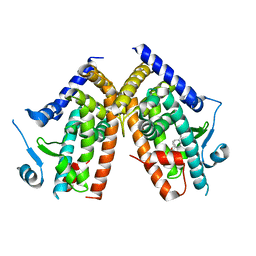 | | Crystal Structure of Human PPAR-gamma Ligand Binding Domain Complexed with an Alpha-Aryloxyphenylacetic Acid Agonist | | Descriptor: | (2S)-(4-ISOPROPYLPHENYL)[(2-METHYL-3-OXO-5,7-DIPROPYL-2,3-DIHYDRO-1,2-BENZISOXAZOL-6-YL)OXY]ACETATE, Peroxisome proliferator activated receptor gamma | | Authors: | Shi, G.Q, Dropinski, J.F, McKeever, B.M, Adams, A.D, MacNaul, K.L, Elbrecht, A, Berger, J.P, Zhou, G, Doebber, T.W. | | Deposit date: | 2005-04-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Synthesis of alpha-Aryloxyphenylacetic Acid Derivatives: A Novel Class of PPAR alpha/gamma Dual Agonists with Potent Antihyperglycemic and Lipid Modulating Activity
J.Med.Chem., 48, 2005
|
|
4E26
 
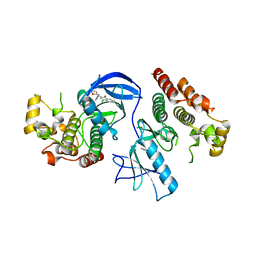 | | BRAF in complex with an organic inhibitor 7898734 | | Descriptor: | 5-chloro-7-[(R)-furan-2-yl(pyridin-2-ylamino)methyl]quinolin-8-ol, Serine/threonine-protein kinase B-raf | | Authors: | Qin, J, Xie, P, Ventocilla, C, Zhou, G, Vultur, A, Chen, Q, Herlyn, M, Winkler, J, Marmorstein, R. | | Deposit date: | 2012-03-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of a Novel Family of BRAF(V600E) Inhibitors.
J.Med.Chem., 55, 2012
|
|
3EDZ
 
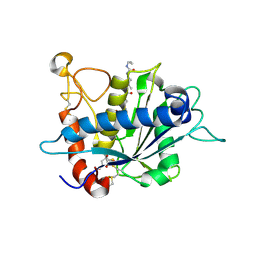 | | Crystal structure of catalytic domain of TACE with hydroxamate inhibitor | | Descriptor: | ADAM 17, CITRIC ACID, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Mazzola, R.D, Zhu, Z, Sinning, L, McKittrick, B, Lavey, B, Spitler, J, Kozlowski, J, Neng-Yang, S, Zhou, G, Guo, Z, Orth, P, Madison, V, Sun, J, Lundell, D, Niu, X. | | Deposit date: | 2008-09-03 | | Release date: | 2008-09-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel hydroxamates as highly potent tumor necrosis factor-alpha converting enzyme inhibitors. Part II: optimization of the S3' pocket.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1D41
 
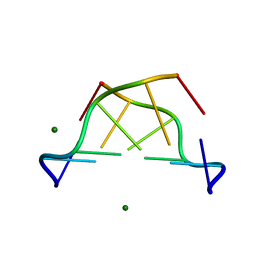 | |
3KME
 
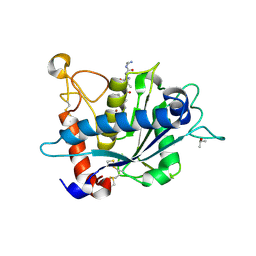 | | Crystal structure of catalytic domain of TACE with phenyl-pyrrolidinyl-tartrate inhibitor | | Descriptor: | (2R,3R)-2,3-dihydroxy-4-oxo-4-[(2R)-2-phenylpyrrolidin-1-yl]-N-(thiophen-2-ylmethyl)butanamide, ISOPROPYL ALCOHOL, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The discovery of novel tartrate-based TNF-alpha converting enzyme (TACE) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2009
|
|
1M15
 
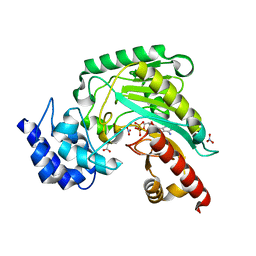 | | Transition state structure of arginine kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, MAGNESIUM ION, ... | | Authors: | Yousef, M.S, Fabiola, F, Gattis, J.L, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2002-06-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Refinement of the arginine kinase transition-state analogue complex at 1.2 A resolution: mechanistic insights.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3KMC
 
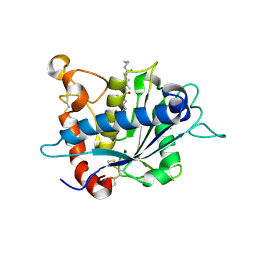 | | Crystal structure of catalytic domain of TACE with tartrate-based inhibitor | | Descriptor: | (2R,3R)-4-[4-(2-chlorophenyl)piperazin-1-yl]-2,3-dihydroxy-4-oxo-N-(2-thiophen-2-ylethyl)butanamide, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, TNF-alpha-converting enzyme, ... | | Authors: | Orth, P. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The discovery of novel tartrate-based TNF-alpha converting enzyme (TACE) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2009
|
|
3L0V
 
 | |
3L0T
 
 | | Crystal structure of catalytic domain of TACE with hydantoin inhibitor | | Descriptor: | Disintegrin and metalloproteinase domain-containing protein 17, ISOPROPYL ALCOHOL, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery and SAR of hydantoin TACE inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LEA
 
 | |
3LGP
 
 | | Crystal structure of catalytic domain of tace with benzimidazolyl-thienyl-tartrate based inhibitor | | Descriptor: | (2R,3R)-4-[(2R)-2-(3-chlorophenyl)pyrrolidin-1-yl]-2,3-dihydroxy-4-oxo-N-[(5-{[2-(trifluoromethyl)-1H-benzimidazol-1-yl]methyl}thiophen-2-yl)methyl]butanamide, Disintegrin and metalloproteinase domain-containing protein 17, ZINC ION | | Authors: | Orth, P. | | Deposit date: | 2010-01-21 | | Release date: | 2010-07-28 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity relationships of tartrate-based TACE inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LE9
 
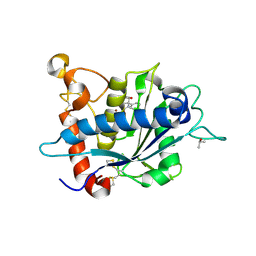 | |
3TY0
 
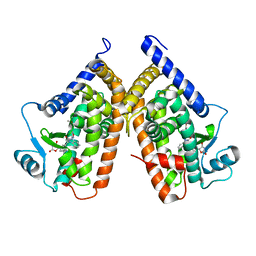 | | Structure of PPARgamma ligand binding domain in complex with (R)-5-(3-((3-(6-methoxybenzo[d]isoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)methyl)phenyl)-5-methyloxazolidine-2,4-dione | | Descriptor: | (5R)-5-(3-{[3-(6-methoxy-1,2-benzoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl]methyl}phenyl)-5-methyl-1,3-oxazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Soisson, S.M, Meinke, P.M, McKeever, B, Liu, W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Benzimidazolones: a new class of selective peroxisome proliferator-activated receptor gamma (PPAR-gamma) modulators.
J.Med.Chem., 54, 2011
|
|
1D40
 
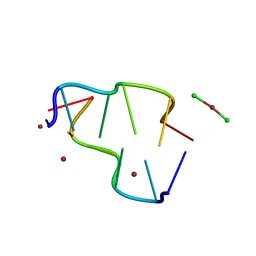 | | BASE SPECIFIC BINDING OF COPPER(II) TO Z-DNA: THE 1.3-ANGSTROMS SINGLE CRYSTAL STRUCTURE OF D(M5CGUAM5CG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) CHLORIDE, COPPER (II) ION, DNA (5'-D(*(5CM)P*(CU)GP*UP*AP*(5CM)P*(CU)G)-3') | | Authors: | Geierstanger, B.H, Kagawa, T.F, Chen, S.-L, Quigley, G.J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Base-specific binding of copper(II) to Z-DNA. The 1.3-A single crystal structure of d(m5CGUAm5CG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
1VHR
 
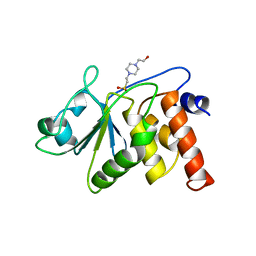 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE VHR, SULFATE ION | | Authors: | Yuvaniyama, J, Denu, J.M, Dixon, J.E, Saper, M.A. | | Deposit date: | 1996-02-20 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the dual specificity protein phosphatase VHR.
Science, 272, 1996
|
|
3E8R
 
 | | Crystal structure of catalytic domain of TACE with hydroxamate inhibitor | | Descriptor: | (1R,2S)-N~2~-hydroxy-1-{4-[(2-phenylquinolin-4-yl)methoxy]benzyl}cyclopropane-1,2-dicarboxamide, ADAM 17, CITRIC ACID, ... | | Authors: | Orth, P. | | Deposit date: | 2008-08-20 | | Release date: | 2008-10-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel hydroxamates as highly potent tumor necrosis factor-alpha converting enzyme inhibitors. Part II: optimization of the S3' pocket.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1SD0
 
 | | Structure of arginine kinase C271A mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Gattis, J.L, Ruben, E, Fenley, M.O, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2004-02-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The active site cysteine of arginine kinase: structural and functional analysis of partially active mutants
Biochemistry, 43, 2004
|
|
6A69
 
 | | Cryo-EM structure of a P-type ATPase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroplastin, Plasma membrane calcium-transporting ATPase 1 | | Authors: | Gong, D.S, Chi, X.M, Ren, K, Huang, G.X.Y, Zhou, G.W, Yan, N, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin.
Nat Commun, 9, 2018
|
|
6JGZ
 
 | | Structure of RyR2 (F/P/Ca2+ dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JG3
 
 | | Cryo-EM structure of RyR2 (Ca2+ alone dataset) | | Descriptor: | Ryanodine receptor 2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JHN
 
 | | Structure of RyR2 (F/C/Ca2+ dataset) | | Descriptor: | CAFFEINE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JH6
 
 | | Structure of RyR2 (F/A/Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1QH4
 
 | | CRYSTAL STRUCTURE OF CHICKEN BRAIN-TYPE CREATINE KINASE AT 1.41 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CALCIUM ION, CREATINE KINASE | | Authors: | Eder, M, Schlattner, U, Becker, A, Wallimann, T, Kabsch, W, Fritz-Wolf, K. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of brain-type creatine kinase at 1.41 A resolution.
Protein Sci., 8, 1999
|
|
