1PLS
 
 | | SOLUTION STRUCTURE OF A PLECKSTRIN HOMOLOGY DOMAIN | | Descriptor: | PLECKSTRIN HOMOLOGY DOMAIN | | Authors: | Yoon, H.S, Hajduk, P.J, Petros, A.M, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1994-05-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pleckstrin-homology domain.
Nature, 369, 1994
|
|
2KI3
 
 | |
5WXE
 
 | | Highly disulfide-constrained antifeedant jasmintides from Jasminum sambac flowers | | Descriptor: | jasmintide js3 | | Authors: | Kumari, G, Wong, K.H, Serra, A, Shin, J, Yoon, H.S, Sze, S.K, James, P.T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Highly disulfide-constrained antifeedant jasmintides from Jasminum sambac flowers
To Be Published
|
|
2OFN
 
 | | Solution structure of FK506-binding domain (FKBD)of FKBP35 from Plasmodium falciparum | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative | | Authors: | Kang, C.B, Ye, H, Yoon, H.R, Yoon, H.S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FK506 binding domain (FKBD) of Plasmodium falciparum FK506 binding protein 35 (PfFKBP35).
Proteins, 70, 2007
|
|
2MF9
 
 | | Solution structure of the N-terminal domain of human FKBP38 (FKBP38NTD) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP8 | | Authors: | Kang, C, Ye, H, Simon, B, Sattler, M, Yoon, H.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional role of the flexible N-terminal extension of FKBP38 in catalysis.
Sci Rep, 3, 2013
|
|
3IHZ
 
 | | Crystal structure of the FK506 binding domain of Plasmodium vivax FKBP35 in complex with FK506 | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Qureshi, I.A, Alag, R, Yoon, H.S, Lescar, J. | | Deposit date: | 2009-07-31 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Protein Sci., 19, 2010
|
|
3PA7
 
 | | Crystal structure of FKBP from plasmodium vivax in complex with tetrapeptide ALPF | | Descriptor: | 4-mer Peptide ALPF, 70 kDa peptidylprolyl isomerase, putative | | Authors: | Balakrishna, A.M, Alag, R, Yoon, H.S. | | Deposit date: | 2010-10-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax
EUKARYOTIC CELL, 12, 2013
|
|
1MAZ
 
 | | X-RAY STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH | | Descriptor: | Bcl-2-like protein 1 | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
1LXL
 
 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
4ITZ
 
 | |
4JYS
 
 | | Crystal structure of FKBP25 from Plasmodium Vivax | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Peptidyl-prolyl cis-trans isomerase | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Plasmodium vivax FK506-binding protein 25 reveals conformational changes responsible for its noncanonical activity
Proteins, 82, 2014
|
|
4J4O
 
 | | Crystal structure of FK506 binding domain of plasmodium VIVAX FKBP35 in complex with D44 | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative, GLYCEROL, ... | | Authors: | Sreekanth, R, Harikishore, A, Yoon, H.S. | | Deposit date: | 2013-02-07 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Small molecule Plasmodium FKBP35 inhibitor as a potential antimalaria agent.
Sci Rep, 3, 2013
|
|
4J4N
 
 | |
1DHM
 
 | | DNA-BINDING DOMAIN OF E2 FROM HUMAN PAPILLOMAVIRUS-31, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2 PROTEIN | | Authors: | Liang, H, Petros, A.P, Meadows, R.P, Yoon, H.S, Egan, D.A, Walter, K, Holzman, T.F, Robins, T, Fesik, S.W. | | Deposit date: | 1995-08-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of a human papillomavirus E2 protein: evidence for flexible DNA-binding regions.
Biochemistry, 35, 1996
|
|
1GJH
 
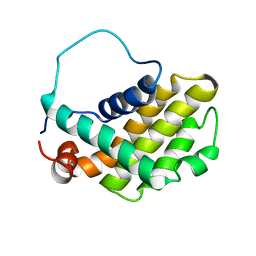 | | HUMAN BCL-2, ISOFORM 2 | | Descriptor: | PROTEIN (APOPTOSIS REGULATOR BCL-2 WITH PUTATIVE FLEXIBLE LOOP REPLACED WITH A PORTION OF APOPTOSIS REGULATOR BCL-X PROTEIN) | | Authors: | Petros, A.M, Medek, A, Nettesheim, D.G, Kim, D.H, Yoon, H.S, Swift, K, Matayoshi, E.D, Oltersdorf, T, Fesik, S.W. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antiapoptotic protein bcl-2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G5M
 
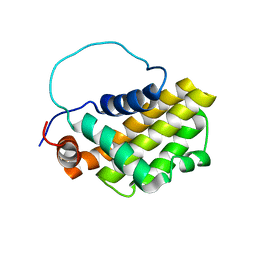 | | HUMAN BCL-2, ISOFORM 1 | | Descriptor: | PROTEIN (APOPTOSIS REGULATOR BCL-2 WITH PUTATIVE FLEXIBLE LOOP REPLACED WITH A PORTION OF APOPTOSIS REGULATOR BCL-X PROTEIN) | | Authors: | Petros, A.M, Medek, A, Nettesheim, D.G, Kim, D.H, Yoon, H.S, Swift, K, Matayoshi, E.D, Oltersdorf, T, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antiapoptotic protein bcl-2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1TCE
 
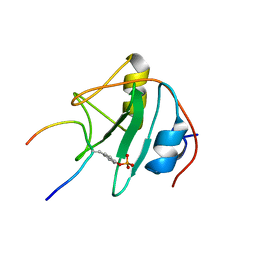 | | SOLUTION NMR STRUCTURE OF THE SHC SH2 DOMAIN COMPLEXED WITH A TYROSINE-PHOSPHORYLATED PEPTIDE FROM THE T-CELL RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PHOSPHOPEPTIDE OF THE ZETA CHAIN OF T CELL RECEPTOR, SHC | | Authors: | Zhou, M.-M, Meadows, R.P, Logan, T.M, Yoon, H.S, Wade, W.R, Ravichandran, K.S, Burakoff, S.J, Feisk, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Shc SH2 domain complexed with a tyrosine-phosphorylated peptide from the T-cell receptor.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2L9L
 
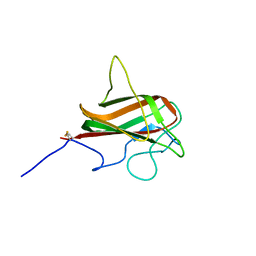 | | NMR Structure of the Mouse MFG-E8 C2 Domain | | Descriptor: | Lactadherin | | Authors: | Ye, H, Yoon, H.S. | | Deposit date: | 2011-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of C2 domain of MFG-E8 and insights into its molecular recognition with phosphatidylserine
Biochim.Biophys.Acta, 1828, 2013
|
|
2LAV
 
 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
3UQI
 
 | |
4MGV
 
 | |
3NI6
 
 | |
4PPI
 
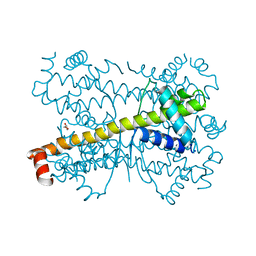 | | Crystal structure of Bcl-xL hexamer | | Descriptor: | Bcl-2-like protein 1, GLYCEROL | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
2VN1
 
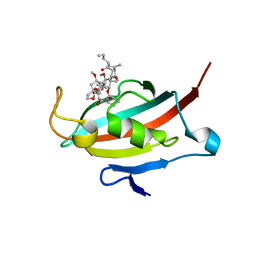 | | Crystal structure of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with FK506 | | Descriptor: | 70 KDA PEPTIDYLPROLYL ISOMERASE, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Kotaka, M, Alag, R, Ye, H, Preiser, P.R, Yoon, H.S, Lescar, J. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Fk506 Binding Domain of Plasmodium Falciparum Fkbp35 in Complex with Fk506.
Biochemistry, 47, 2008
|
|
4QVF
 
 | |
