1UAW
 
 | | Solution structure of the N-terminal RNA-binding domain of mouse Musashi1 | | Descriptor: | mouse-musashi-1 | | Authors: | Miyanoiri, Y, Kobayashi, H, Watanabe, M, Ikeda, T, Nagata, T, Okano, H, Uesugi, S, Katahira, M. | | Deposit date: | 2003-03-24 | | Release date: | 2004-03-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics
J.Biol.Chem., 278, 2003
|
|
2KIA
 
 | | Solution structure of Myosin VI C-terminal cargo-binding domain | | Descriptor: | Myosin-VI | | Authors: | Feng, W, Yu, C, Wei, Z, Miyanoiri, Y, Zhang, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Myosin VI undergoes cargo-mediated dimerization
Cell(Cambridge,Mass.), 138, 2009
|
|
3WIY
 
 | | Crystal structure of Mcl-1 in complex with compound 10 | | Descriptor: | 7-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]carbamoyl}-2-methylphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Sogabe, S, Igaki, S, Hayano, Y. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
1IQT
 
 | | Solution structure of the C-terminal RNA-binding domain of heterogeneous nuclear ribonucleoprotein D0 (AUF1) | | Descriptor: | heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Katahira, M, Miyanoiri, Y, Enokizono, Y, Matsuda, G, Nagata, T, Ishikawa, F, Uesugi, S. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal RNA-binding domain of hnRNP D0 (AUF1), its interactions with RNA and DNA, and change in backbone dynamics upon complex formation with DNA.
J.Mol.Biol., 311, 2001
|
|
3WIZ
 
 | | Crystal structure of Bcl-xL in complex with compound 10 | | Descriptor: | 7-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]carbamoyl}-2-methylphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Bcl-2-like protein 1, PHOSPHATE ION | | Authors: | Sogabe, S, Igaki, S, Hayano, Y. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
3WIX
 
 | | Crystal structure of Mcl-1 in complex with compound 4 | | Descriptor: | 7-(4-carboxyphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Sogabe, S, Igaki, S, Hayano, Y. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
7E0D
 
 | | Structure of L-glutamate oxidase R305E mutant in complex with L-arginine | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
7E0C
 
 | | Structure of L-glutamate oxidase R305E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1F3P
 
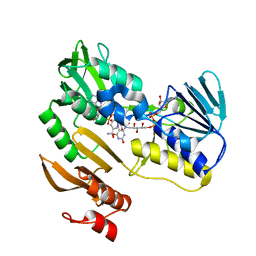 | | FERREDOXIN REDUCTASE (BPHA4)-NADH COMPLEX | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsuidagger, Y. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
8K9O
 
 | |
7SQO
 
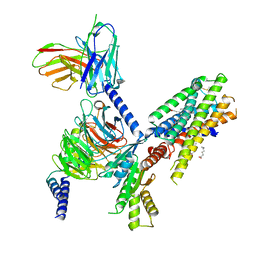 | | Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | McGrath, A.P, Kang, Y, Flinspach, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
2JMW
 
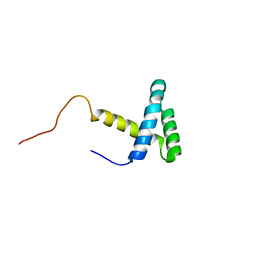 | | Structure of DNA-Binding Domain of Arabidopsis GT-1 | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Niyada, E, Noto, K, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
7SR8
 
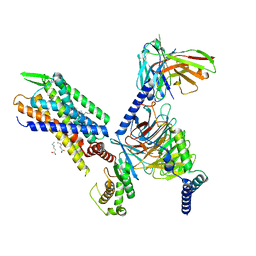 | | Molecular mechanism of the the wake-promoting agent TAK-925 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hypocretin receptor type 2, ... | | Authors: | Yin, J, Chapman, K, Lian, P, De Brabander, J.K, Rosenbaum, D.M. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
3H8D
 
 | | Crystal structure of Myosin VI in complex with Dab2 peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Disabled homolog 2, ... | | Authors: | Yu, C, Feng, W, Wei, Z, Zhang, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Myosin VI undergoes cargo-mediated dimerization
Cell(Cambridge,Mass.), 138, 2009
|
|
1X0F
 
 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D(AUF1) with telomeric DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D.
J.Biol.Chem., 280, 2005
|
|
1WTB
 
 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D (AUF1) with telomere DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2004-11-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D
J.Biol.Chem., 280, 2005
|
|
5B4O
 
 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | Authors: | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
2EBI
 
 | | Arabidopsis GT-1 DNA-binding domain with T133D phosphomimetic mutation | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Noto, K, Niyada, E, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2007-02-08 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
7CKV
 
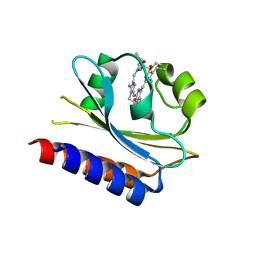 | | Crystal structure of Cyanobacteriochrome GAF domain in Pr state | | Descriptor: | MAGNESIUM ION, PHYCOCYANOBILIN, RcaE | | Authors: | Nagae, T, Koizumi, T, Hirose, Y, Mishima, M. | | Deposit date: | 2020-07-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of the protochromic green/red photocycle of the chromatic acclimation sensor RcaE.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3VVI
 
 | |
3WYM
 
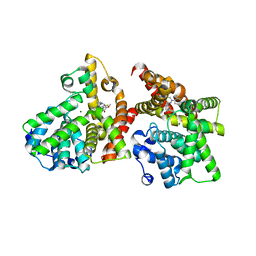 | | Crystal structure of the catalytic domain of PDE10A complexed with 1-(2-fluoro-4-(1H-pyrazol-1-yl)phenyl)-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Hayano, Y. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor.
J.Med.Chem., 57, 2014
|
|
3WYK
 
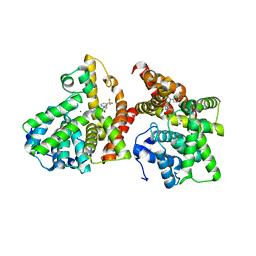 | | Crystal structure of the catalytic domain of PDE10A complexed with 3-(1-phenyl-1H-pyrazol-5-yl)-1-(3-(trifluoromethyl)phenyl)pyridazin-4(1H)-one | | Descriptor: | 3-(1-phenyl-1H-pyrazol-5-yl)-1-[3-(trifluoromethyl)phenyl]pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Hayano, Y. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor.
J.Med.Chem., 57, 2014
|
|
3WYL
 
 | | Crystal structure of the catalytic domain of PDE10A complexed with 5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)-1-(3-(trifluoromethyl)phenyl)pyridazin-4(1H)-one | | Descriptor: | 5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)-1-[3-(trifluoromethyl)phenyl]pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Hayano, Y. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor.
J.Med.Chem., 57, 2014
|
|
8HN3
 
 | | Soluble domain of cytochrome c-556 from Chlorobaculum tepidum | | Descriptor: | ACETATE ION, Cytochrome c-556, GLYCEROL, ... | | Authors: | Kishimoto, H, Azai, C, Yamamoto, T, Mutoh, R, Nakaniwa, T, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
