2F2F
 
 | | Crystal structure of cytolethal distending toxin (CDT) from Actinobacillus actinomycetemcomitans | | Descriptor: | Cytolethal distending toxin A, Cytolethal distending toxin B, cytolethal distending toxin C | | Authors: | Yamada, T, Komoto, J, Saiki, K, Konishi, K, Takusagawa, F. | | Deposit date: | 2005-11-16 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Variation of loop sequence alters stability of cytolethal distending toxin (CDT): crystal structure of CDT from Actinobacillus actinomycetemcomitans
Protein Sci., 15, 2006
|
|
2RKB
 
 | | Serine dehydratase like-1 from human cancer cells | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine dehydratase-like | | Authors: | Yamada, T, Komoto, J, Kasuya, T, Mori, H, Ogawa, H, Takusagawa, F. | | Deposit date: | 2007-10-16 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A catalytic mechanism that explains a low catalytic activity of serine dehydratase like-1 from human cancer cells: Crystal structure and site-directed mutagenesis studies.
Biochim.Biophys.Acta, 1780, 2008
|
|
2H5L
 
 | | S-Adenosylhomocysteine hydrolase containing NAD and 3-deaza-D-eritadenine | | Descriptor: | (2R,3R)-4-(4-AMINO-1H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-2,3-DIHYDROXYBUTANOIC ACID, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, T, Komoto, J, Takusagawa, F. | | Deposit date: | 2006-05-26 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of eritadenine and its 3-deaza analogues: Potent inhibitors of S-adenosylhomocysteine hydrolase and hypocholesterolemic agents.
Biochem.Pharm., 73, 2007
|
|
1XWF
 
 | | K185N mutated S-adenosylhomocysteine hydrolase | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, T, Takata, Y, Komoto, J, Gomi, T, Ogawa, H, Fujioka, M, Takusagawa, F. | | Deposit date: | 2004-11-01 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanism of S-adenosylhomocysteine hydrolase: Roles of His 54, Asp130, Glu155, Lys185, and Aspl89.
Int.J.Biochem.Cell Biol., 37, 2005
|
|
1PWH
 
 | | Rat Liver L-Serine Dehydratase- Complex with PYRIDOXYL-(O-METHYL-SERINE)-5-MONOPHOSPHATE | | Descriptor: | L-serine dehydratase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-O-METHYL-L-SERINE, POTASSIUM ION | | Authors: | Yamada, T, Komoto, J, Takata, Y, Ogawa, H, Takusagawa, F. | | Deposit date: | 2003-07-01 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of serine dehydratase from rat liver.
Biochemistry, 42, 2003
|
|
1PWE
 
 | | Rat Liver L-Serine Dehydratase Apo Enzyme | | Descriptor: | L-serine dehydratase | | Authors: | Yamada, T, Komoto, J, Takata, Y, Ogawa, H, Takusagawa, F. | | Deposit date: | 2003-07-01 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of serine dehydratase from rat liver.
Biochemistry, 42, 2003
|
|
6KK8
 
 | | XN joint refinement of manganese catalase from Thermus Thermophilus HB27 | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (III) ION, OXYGEN ATOM, ... | | Authors: | Yamada, T, Yano, N, Kusaka, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.37 Å), X-RAY DIFFRACTION | | Cite: | Single-crystal time-of-flight neutron Laue methods: application to manganese catalase from Thermus thermophilus HB27
J.Appl.Crystallogr., 2019
|
|
1Z9H
 
 | | Microsomal prostaglandin E synthase type-2 | | Descriptor: | ACETATE ION, CHLORIDE ION, INDOMETHACIN, ... | | Authors: | Yamada, T, Komoto, J, Watanabe, K, Ohmiya, Y, Takusagawa, F. | | Deposit date: | 2005-04-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Possible Catalytic Mechanism of Microsomal Prostaglandin E Synthase Type 2 (mPGES-2).
J.Mol.Biol., 348, 2005
|
|
3VXF
 
 | | X/N Joint refinement of Human alpha-thrombin-Bivalirudin complex PD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.602 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
3VXE
 
 | | Human alpha-thrombin-Bivalirudin complex at PD5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
1LZ5
 
 | | STRUCTURAL AND FUNCTIONAL ANALYSES OF THE ARG-GLY-ASP SEQUENCE INTRODUCED INTO HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, HUMAN LYSOZYME | | Authors: | Matsushima, M, Inaka, K, Yamada, T, Sekiguchi, K, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme.
J.Biol.Chem., 268, 1993
|
|
7YVQ
 
 | | Complex structure of Clostridioides difficile binary toxin folded CDTa-bound CDTb-pore (short). | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7YVS
 
 | | Complex structure of Clostridioides difficile binary toxin unfolded CDTa-bound CDTb-pore (short). | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7YVW
 
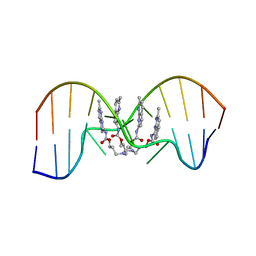 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
1LZ6
 
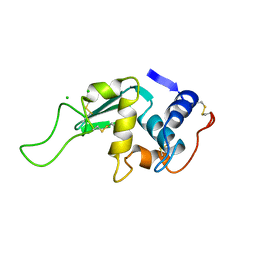 | | STRUCTURAL AND FUNCTIONAL ANALYSES OF THE ARG-GLY-ASP SEQUENCE INTRODUCED INTO HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, HUMAN LYSOZYME | | Authors: | Matsushima, M, Inaka, K, Yamada, T, Sekiguchi, K, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme.
J.Biol.Chem., 268, 1993
|
|
2PBJ
 
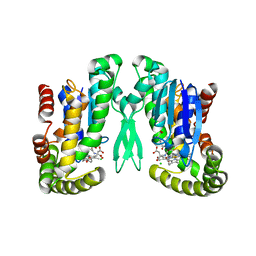 | | GSH-heme bound microsomal prostaglandin E synthase | | Descriptor: | CHLORIDE ION, GLUTATHIONE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Takusagawa, F, Yamada, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | PGH2 degradation pathway catalyzed by GSH-heme complex bound microsomal prostaglandin E2 synthase type 2: the first example of a dual-function enzyme.
Biochemistry, 46, 2007
|
|
7VNN
 
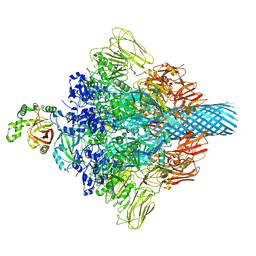 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, CALCIUM ION, CdtA | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7VNJ
 
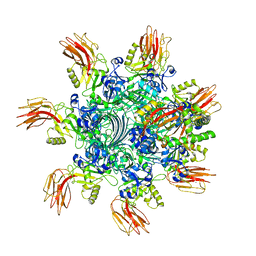 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosyltransferase enzymatic component, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
6KLW
 
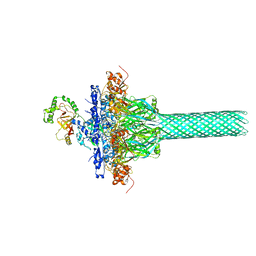 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLX
 
 | | Pore structure of Iota toxin binding component (Ib) | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLO
 
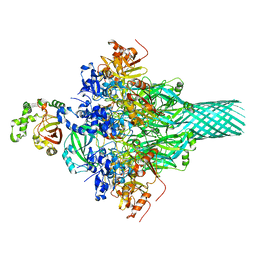 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3U2J
 
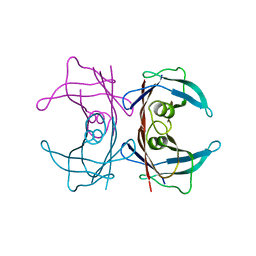 | | Neutron crystal structure of human Transthyretin | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3U2I
 
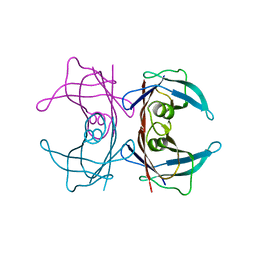 | | X-ray crystal structure of human Transthyretin at room temperature | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
7OZ4
 
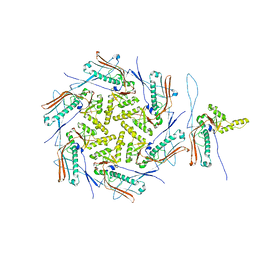 | | Mature capsid of bacteriophage phiRSA1 | | Descriptor: | p2 family phage major capsid protein | | Authors: | Effantin, G, Fujiwara, A, Kawsaki, T, Yamada, T, Schoehn, G. | | Deposit date: | 2021-06-25 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High Resolution Structure of the Mature Capsid of Ralstonia solanacearum Bacteriophage phi RSA1 by Cryo-Electron Microscopy.
Int J Mol Sci, 22, 2021
|
|
2F38
 
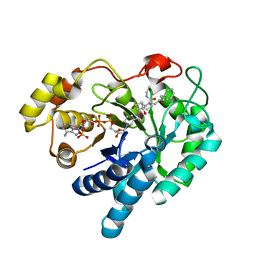 | | Crystal structure of prostaglandin F synathase containing bimatoprost | | Descriptor: | (5Z)-7-{(1R,2R,3R,5S)-3,5-DIHYDROXY-2-[(1E,3S)-3-HYDROXY-5-PHENYLPENT-1-ENYL]CYCLOPENTYL}-N-ETHYLHEPT-5-ENAMIDE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Komoto, J, Yamada, T, Watanabe, K, Woodward, D.F, Takusagawa, F. | | Deposit date: | 2005-11-18 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prostaglandin F2alpha formation from prostaglandin H2 by prostaglandin F synthase (PGFS): crystal structure of PGFS containing bimatoprost.
Biochemistry, 45, 2006
|
|
