7B8R
 
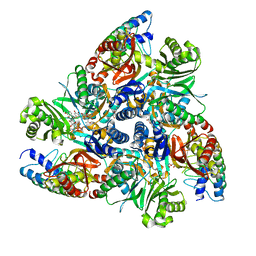 | | Doxycycline bound structure of bacterial efflux pump. | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, DARPin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilhelm, J, Sjuts, H, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7B8S
 
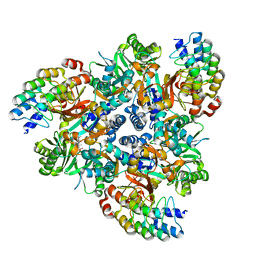 | | Fusidic acid bound structure of bacterial efflux pump. | | Descriptor: | DARPin, FUSIDIC ACID, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Wilhelm, J, Sjuts, H, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7B8T
 
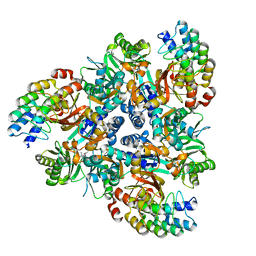 | | Levofloxacin bound structure of bacterial efflux pump. | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Wilhelm, J, Sjuts, H, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
2K2G
 
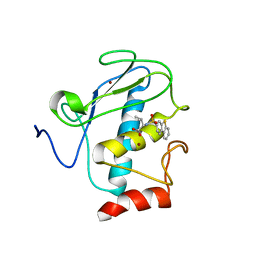 | | Solution structure of the wild-type catalytic domain of human matrix metalloproteinase 12 (MMP-12) in complex with a tight-binding inhibitor | | Descriptor: | Macrophage metalloelastase, N-(dibenzo[b,d]thiophen-3-ylsulfonyl)-L-valine, ZINC ION | | Authors: | Markus, M.A, Dwyer, B, Wolfrom, S, Li, J, Li, W, Malakian, K, Wilhelm, J, Tsao, D.H.H. | | Deposit date: | 2008-04-01 | | Release date: | 2008-05-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of wild-type human matrix metalloproteinase 12 (MMP-12) in complex with a tight-binding inhibitor.
J.Biomol.Nmr, 41, 2008
|
|
1IJZ
 
 | |
1IK0
 
 | |
6Y7B
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-carbopyronine fluorophore substrate | | Descriptor: | 4-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]-2-[3-(dimethylamino)-6-(dimethyl-$l^{4}-azanylidene)-10,10-dimethyl-anthracen-9-yl]benzoic acid, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
6Y7A
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
6Y8P
 
 | | Crystal structure of SNAP-tag labeled with a benzyl-tetramethylrhodamine fluorophore | | Descriptor: | 1,2-ETHANEDIOL, O6-alkylguanine-DNA alkyltransferase mutant, ZINC ION, ... | | Authors: | Gotthard, G, Tanzer, T, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
6ZCC
 
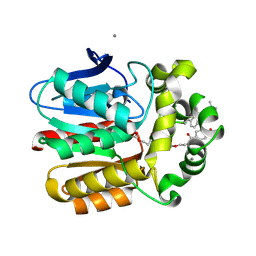 | | X-ray structure of the Haloalkane dehalogenase HOB (HaloTag7-based Oligonucleotide Binder) labeled with a chloroalkane-tetramethylrhodamine fluorophore substrate | | Descriptor: | ACETATE ION, CALCIUM ION, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
1AYK
 
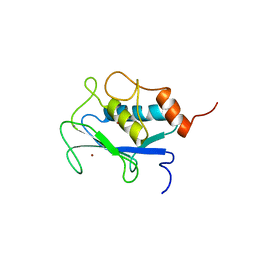 | |
3AYK
 
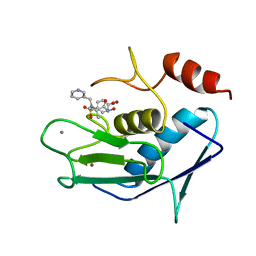 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | Authors: | Powers, R, Moy, F.J. | | Deposit date: | 1999-02-01 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
4AYK
 
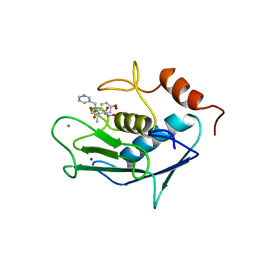 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | Authors: | Powers, R, Moy, F.J. | | Deposit date: | 1999-02-01 | | Release date: | 1999-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
2AYK
 
 | |
8B6N
 
 | |
8B6P
 
 | |
3II5
 
 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | Authors: | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7B8P
 
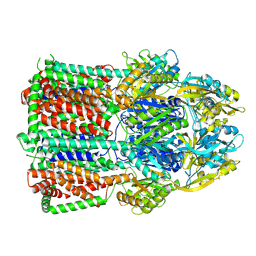 | | Acinetobacter baumannii multidrug transporter AdeB in OOO state | | Descriptor: | Efflux pump membrane transporter | | Authors: | Ornik-Cha, A, Reitz, J, Seybert, A, Frangakis, A, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7B8Q
 
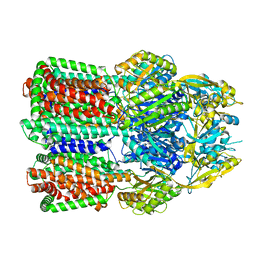 | | Acinetobacter baumannii multidrug transporter AdeB in L*OO state | | Descriptor: | Efflux pump membrane transporter | | Authors: | Ornik-Cha, A, Reitz, J, Seybert, A, Frangakis, A, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7ZIY
 
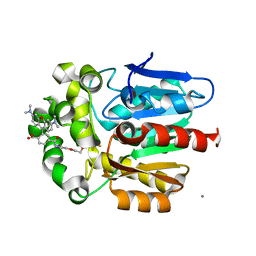 | | X-ray structure of the haloalkane dehalogenase HaloTag7 bound to a pentyltrifluoromethanesulfonamide tetramethylrhodamine ligand (TMR-T5) | | Descriptor: | CALCIUM ION, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-[5-(trifluoromethylsulfonylamino)pentoxy]ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7ZIZ
 
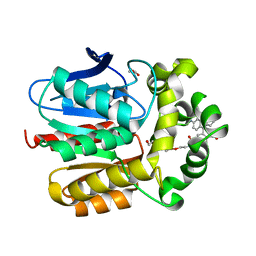 | | X-ray structure of the dead variant haloalkane dehalogenase HaloTag7-D106A bound to a pentanol tetramethylrhodamine ligand (TMR-Hy5) | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7ZJ0
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 bound to a pentylmethanesulfonamide tetramethylrhodamine ligand (TMR-S5) | | Descriptor: | GLYCEROL, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-[5-(methylsulfonylamino)pentoxy]ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
2ID5
 
 | | Crystal Structure of the Lingo-1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine rich repeat neuronal 6A, ... | | Authors: | Mosyak, L, Wood, A, Dwyer, B, Johnson, M, Stahl, M.L, Somers, W.S. | | Deposit date: | 2006-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition.
J.Biol.Chem., 281, 2006
|
|
