7XYB
 
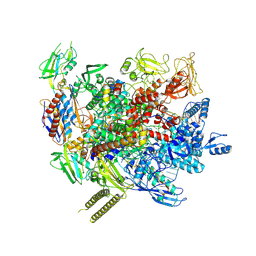 | | The cryo-EM structure of an AlpA-loaded complex | | Descriptor: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wen, A, Feng, Y. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
7XYA
 
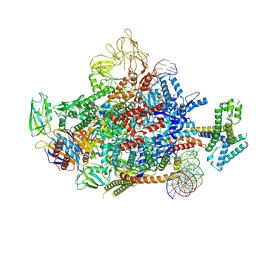 | | The cryo-EM structure of an AlpA-loading complex | | Descriptor: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wen, A, Feng, Y. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
8IGR
 
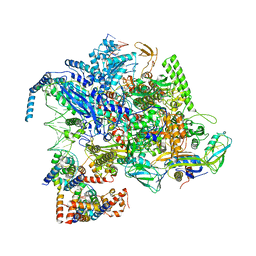 | | Cryo-EM structure of CII-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
8IGS
 
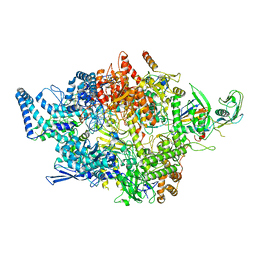 | | Cryo-EM structure of RNAP-promoter open complex at lambda promoter PRE | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
6K4Y
 
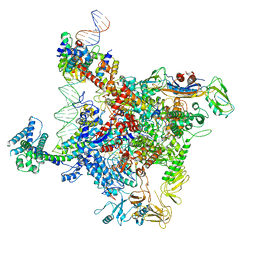 | | CryoEM structure of sigma appropriation complex | | Descriptor: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
6M6B
 
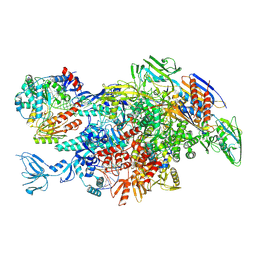 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6A
 
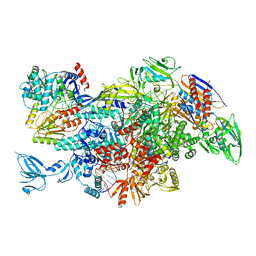 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6LS9
 
 | | Crystal structure of bovine herpesvirus 1 glycoprotein D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6LSA
 
 | | Complex structure of bovine herpesvirus 1 glycoprotein D and bovine nectin-1 IgV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6M6C
 
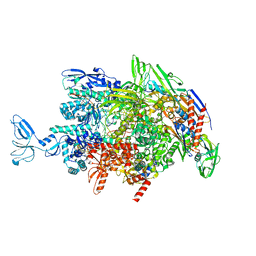 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
7D7D
 
 | | CryoEM structure of gp45-dependent transcription activation complex | | Descriptor: | DNA (nontemplate strand), DNA (template strand), DNA polymerase clamp, ... | | Authors: | Shi, J, Wen, A, Jin, S, Feng, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Transcription activation by a sliding clamp.
Nat Commun, 12, 2021
|
|
7D7C
 
 | | CryoEM structure of gp55-dependent RNA polymerase-promoter open complex | | Descriptor: | DNA (nontemplate strand), DNA (template strand), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Jin, S, Feng, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transcription activation by a sliding clamp.
Nat Commun, 12, 2021
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
8KHQ
 
 | | Bifunctional sulfoxide synthase OvoA_Th2 in complex with histidine and cysteine | | Descriptor: | 5-histidylcysteine sulfoxide synthase/putative 4-mercaptohistidine N1-methyltranferase, COBALT (II) ION, CYSTEINE, ... | | Authors: | Wang, J, Ye, K, Wang, X.Y, Yan, W.P. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Biochemical and Structural Characterization of OvoA Th2 : A Mononuclear Nonheme Iron Enzyme from Hydrogenimonas thermophila for Ovothiol Biosynthesis.
Acs Catalysis, 13, 2023
|
|
7F75
 
 | | Cryo-EM structure of Spx-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of transcription activation by the global regulator Spx.
Nucleic Acids Res., 49, 2021
|
|
7VF9
 
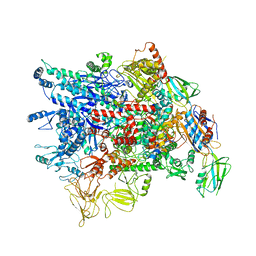 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7VWY
 
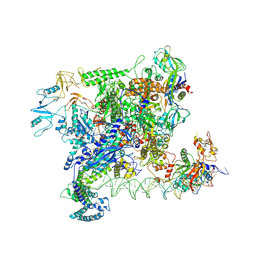 | |
7VWZ
 
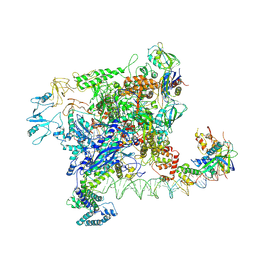 | | Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-12 | | Release date: | 2022-06-08 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator.
Nucleic Acids Res., 50, 2022
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
7F0R
 
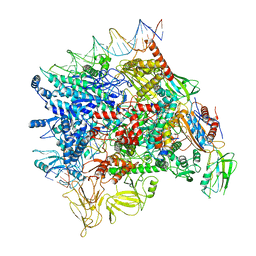 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
6JNX
 
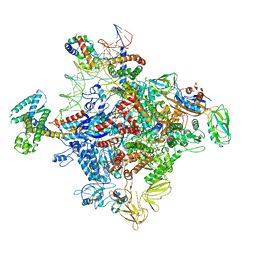 | | Cryo-EM structure of a Q-engaged arrested complex | | Descriptor: | Antiterminator Q protein, DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Feng, Y, Shi, J. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of Q-dependent transcription antitermination.
Nat Commun, 10, 2019
|
|
8GMS
 
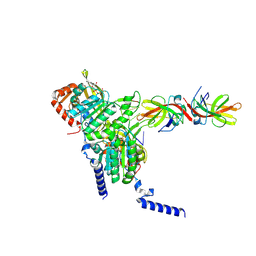 | | Structure of LexA in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), LexA repressor, MAGNESIUM ION, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GMU
 
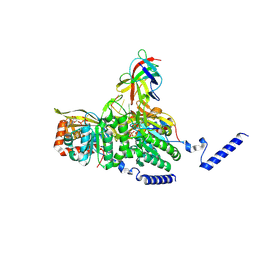 | | Structure of lambda repressor in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GMT
 
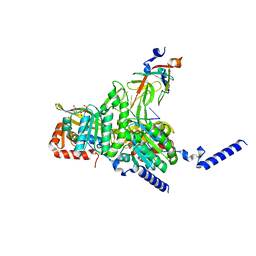 | | Structure of UmuD in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA polymerase V subunit UmuD, MAGNESIUM ION, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6JNY
 
 | |
