5K7U
 
 | |
5K7M
 
 | |
6L42
 
 | |
5K7W
 
 | |
6IUV
 
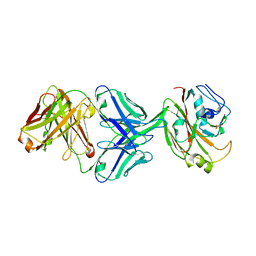 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 3C11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3C11 Heavy Chain, 3C11 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Structural and functional definition of a vulnerable site on the hemagglutinin of highly pathogenic avian influenza A virus H5N1.
J. Biol. Chem., 294, 2019
|
|
4XRU
 
 | | Structure of Pnkp1/Rnl/Hen1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, P. | | Deposit date: | 2015-01-21 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Reconstitution and structure of a bacterial Pnkp1-Rnl-Hen1 RNA repair complex.
Nat Commun, 6, 2015
|
|
8WDK
 
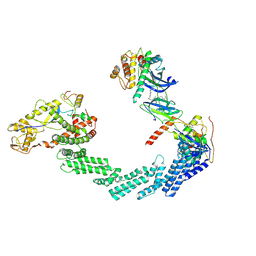 | | The complex structure of Cul2-VCB-Protac-Wee1 | | Descriptor: | (2S,4R)-1-[(2S)-3,3-dimethyl-2-[3-[4-[4-[4-[[3-oxidanylidene-1-[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]-2-prop-2-enyl-pyrazolo[3,4-d]pyrimidin-6-yl]amino]phenyl]piperazin-1-yl]butoxy]propanoylamino]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Wang, P, Zhang, T.T. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of Cul2-VCB-Protac-Wee1 complex at 3.6 Angstrom resolution.
To Be Published
|
|
6IUT
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody AVFluIgG01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, AVFluIgG01 Heavy Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional definition of a vulnerable site on the hemagglutinin of highly pathogenic avian influenza A virus H5N1.
J. Biol. Chem., 294, 2019
|
|
6CKU
 
 | | Solution structure of the zebrafish granulin AaE | | Descriptor: | Granulin-AaE | | Authors: | Wang, P, Ni, F. | | Deposit date: | 2018-02-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of zebrafish progranulins identifies a well-folded granulin/epithelin module protein with pro-cell survival activities.
Protein Sci., 27, 2018
|
|
9F0H
 
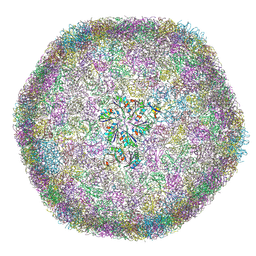 | | cryo-EM structure of carboxysomal mini-shell icosahedral assembly from co-expression of CsoS1C, CsoS4A, and CsoS2-C (T = 9) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell protein CsoS1C, Carboxysome shell vertex protein CsoS4A, ... | | Authors: | Wang, P, Marles-Wright, J, Liu, L.N. | | Deposit date: | 2024-04-16 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
8YVC
 
 | | Cryo-EM structure of carboxysomal midi-shell:icosahedral assembly from CsoS4A/4B/1A/1B/1C/1D and CsoS2 C-terminal co-expression (T = 19) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Wang, P, Li, J.X, Li, T.P, Li, K, Ng, P.C, Wang, S.M, Chriscoli, V, Basle, A, Marles-Wright, J, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
8YVF
 
 | | cryo-EM structure of carboxysomal midi-shell: assembly from CsoS4A/4B/1A/1B/1C/1D and CsoS2 C-terminal co-expression (T=9 Q=12) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Wang, P, Li, J.X, Li, T.P, Li, K, Ng, P.C, Wang, S.M, Chriscoli, V, Basle, A, Marles-Wright, J, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
8YVI
 
 | | Cryo-EM structure of carboxysomal midi-shell: icosahedral assembly from CsoS4A/4B/1A/1B/1C/1D and CsoS2 C-terminal co-expression (T = 13) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Wang, P, Li, J.X, Li, T.P, Li, K, Ng, P.C, Wang, S.M, Chriscoli, V, Basle, A, Marles-Wright, J, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
8YVE
 
 | | cryo-EM structure of carboxysomal midi-shell: icosahedral assembly from CsoS4A/4B/1A/1B/1C/1D and CsoS2 C-terminal co-expression (T = 9) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Wang, P, Li, J.X, Li, T.P, Li, K, Ng, P.C, Wang, S.M, Chriscoli, V, Basle, A, Marles-Wright, J, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
8YXU
 
 | | Crystal structure of CsoS1A/B (modeled with CsoS1A) | | Descriptor: | Major carboxysome shell protein CsoS1A | | Authors: | Wang, P, Li, J.X, Li, T.P, Li, K, Ng, P.C, Wang, S.M, Chriscoli, V, Basle, A, Marles-Wright, J, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2024-04-03 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
8YVD
 
 | | Cryo-EM structure of carboxysomal midi-shell: icosahedral assembly from CsoS4A/4B/1A/1B/1C/1D and CsoS2 C-terminal co-expression (T = 16) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Wang, P, Li, J.X, Li, T.P, Li, K, Ng, P.C, Wang, S.M, Chriscoli, V, Basle, A, Marles-Wright, J, Zhang, Y.Z. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
4QG6
 
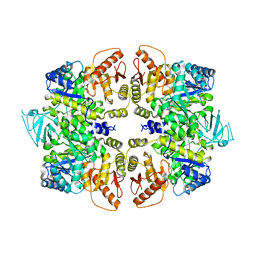 | | crystal structure of PKM2-Y105E mutant | | Descriptor: | PROLINE, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
4QG8
 
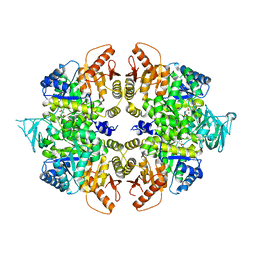 | | crystal structure of PKM2-K305Q mutant | | Descriptor: | GLYCEROL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
5V1D
 
 | |
4QG9
 
 | | crystal structure of PKM2-R399E mutant | | Descriptor: | ACETATE ION, MAGNESIUM ION, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
5SV7
 
 | | The Crystal structure of a chaperone | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wang, P, Li, J, Sha, B. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | The ER stress sensor PERK luminal domain functions as a molecular chaperone to interact with misfolded proteins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4K2X
 
 | |
4RPP
 
 | | crystal structure of PKM2-K422R mutant bound with FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
5U2U
 
 | |
4QGC
 
 | | crystal structure of PKM2-K422R mutant | | Descriptor: | GLYCEROL, POTASSIUM ION, Pyruvate kinase PKM, ... | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
